- Chromatin structure remodeling (RSC) complex
-
RSC is a 17-subunit complex with the capacity to remodel the structure of chromatin. It exhibits a DNA-dependent ATPase activity stimulated by both free and nucleosomal DNA and a capacity to perturb nucleosome structures. It is at least 10-fold more abundant than the SWI/SNF complex and is essential for mitotic growth.[1]
Contents
Generation of loops in dsDNA
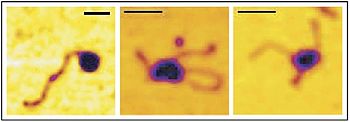
A single molecule study using magnetic tweezers has observed that RSC generates DNA loops in vitro while simultaneously generating negative supercoils in the template.[2] The figure above shows three AFM images from that study. On the left, a single RSC complex is bound to the end of a dsDNA template. The center image shows a DNA loop generated by RSC. The image on the right shows a loop generated by RSC, which contains supercoils.
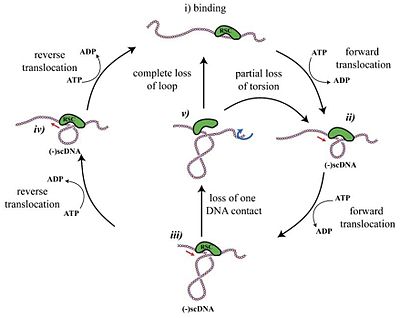
The current model of translocation on dsDNA is shown in the figure to the right, whereby RSC binds the DNA at two locations. Hydrolysis of ATP allows the complex to translocate the DNA into a loop. RSC can release the loop either by translocating back to the original state at a comparable velocity, or by losing one of its two contacts.
RSC components
The following is list of RSC components that have been identified in yeast and the corresponding human orthologs:
yeast human function RSC1 RSC2 RSC3 RSC4 RSC6 SMARCD1, SMARCD2, SMARCD3 RSC7 RSC8 SMARCC1, SMARCC2 RSC9 RSC11 RSC12 RSC14 RSC30 RSC58 See also
- SWI/SNF (nucleosome remodeling complex)
- Transcription coregulator
- Imitation SWI (nucleosome remodeling complex)
References
- ^ Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD (1996). "RSC, an essential, abundant chromatin-remodeling complex". Cell 87 (7): 1249–60. doi:10.1016/S0092-8674(00)81820-6. PMID 8980231.
- ^ Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T (2006). "Direct observation of DNA distortion by the RSC complex". Mol. Cell 21 (3): 417–25. doi:10.1016/j.molcel.2005.12.013. PMID 16455496.
Transcription coregulators Coactivators Corepressors ATP-dependent remodeling factors see also transcription factor/coregulator deficiencies
B bsyn: dna (repl, cycl, reco, repr) · tscr (fact, tcrg, nucl, rnat, rept, ptts) · tltn (risu, pttl, nexn) · dnab, rnab/runp · stru (domn, 1°, 2°, 3°, 4°)External links
This enzyme-related article is a stub. You can help Wikipedia by expanding it.