- Chromatin bridge
-
Chromatin bridge Classification and external resources 
DAPI staining allows for visualization of deoxyribonucleic acid portions of the two daughter cells. The thin “string-like” DNA connecting them is defined as a chromatin bridge.Chromatin bridge is a mitotic occurrence that forms when telomeres of sister chromatids fuse together and fail to completely segregate into their respective daughter cells. Because this event is most prevalent during anaphase, the term anaphase bridge is often used as a substitute. After the formation of individual daughter cells, the DNA bridge connecting homologous chromosomes remains fixed. As the daughter cells exit mitosis and re-enter interphase, the chromatin bridge becomes known as an interphase bridge. These phenomenon are usually visualized using the laboratory techniques of staining and fluorescence microscopy.[1] [2]
Contents
Background
The faithful inheritance of genetic information from one cellular generation to the next heavily relies on the duplication of deoxyribonucleic acid (DNA), as well as the formation of two identical daughter cells. This complicated cellular process, known as mitosis, depends on a multitude of cellular checkpoints, signals, interactions and signal cascades for accurate and faithful functioning. Cancer, characterized by uncontrollable cell growth mechanisms and high tendencies for proliferation and metastasis, is highly prone to mitotic mistakes. As a result, several forms of chromosomal aberrations occur, including, but not limited to, binucleated cells, multipolar spindles and micronuclei.[3] Chromatin bridges may serve as a marker of cancer activity.
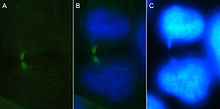 A. Microtubules localized at a chromatin bridge. These polymers are stained with anti-tubulin antibody and viewed using fluorescence microscopy. B. Merged images of two daughter cells connected by a chromatin bridge. The fluorescence techniques of indirect immunofluorescence and DAPI staining were utilized. C. The same cells visualized using DAPI staining.
A. Microtubules localized at a chromatin bridge. These polymers are stained with anti-tubulin antibody and viewed using fluorescence microscopy. B. Merged images of two daughter cells connected by a chromatin bridge. The fluorescence techniques of indirect immunofluorescence and DAPI staining were utilized. C. The same cells visualized using DAPI staining.
Process of Formation
Telomeres are repetitive oligonucleotide sequences of DNA present at the end of each chromosome. They typically form loop structures, called telomere loops, or t-loops. A complex of binding proteins facilitate in ensuring the T-loop is correctly folded during and after each round of replication. Telomere shortening occurs after each mitotic cycle, as a result of DNA polymerase’s inability to copy DNA from the 5’ end of the lagging strand. The mitotic division of normal cells is partially controlled by the Hayflick limit, the total number of times division may occur before the critical length of the telomeres is reached, forcing cells to stop proliferation and undergo senescence.[4] Cancer cells possess specific mechanisms to evade this process. However, the telomerase activity of cancer cells is not extremely efficient, and errors may lead to telomeric shortening. Consequently, T-loops will not form due to the shortening of the telomeres past the point where it can fold back onto itself.[5] In addition, the loss of activity of any cofactor or protein in the protein complex may result in the loss of T-loops. Chromatin bridges are prone to break due to the increased tension resulting from daughter cells moving apart during mitosis.[6] The tension created by this event may lead to either deletion or amplification of genes. It has previously been observed that this phenomenon will repeat in subsequent cell divisions. This cycle of chromatin bridge breakage and fusion ultimately lead to additional chromosomal changes and further instability.[7]
Homologous chromosomes of a dividing, mitotic cell contain complementary telomeric sequences. This implies that two telomeres from homologous chromosomes have the potential to bind via Watson-Crick base pairing, as one strand possesses the complement of the other strand. Adenine binds to thymine and cytosine binds to guanine. In the instance listed below, the nucleotides observed in Strand 2 are complementary to those in Strand 1.
Strand 1 ---A G T T A C T A G C ---
Strand 2 ---T C A A T G A T C G ---
During instances when T-loops do not form, the complementary nature of the telomeric ends results in nucleic acid hybridization as a result of their sticky ends. This leads to homologous chromosomal telomeres fusing together to form a chromatin bridge.Fluorescence Techniques
 A chromatin bridge, visualized using DAPI staining.
A chromatin bridge, visualized using DAPI staining.
Chromatin bridges can be viewed utilizing a laboratory technique known as fluorescence microscopy. Fluorescence is the process that involves excitation of a fluorophore (a molecule with the ability to emit fluorescent light in the visible light spectrum) using ultraviolet light. After the fluorophore becomes chemically excited by the presence of UV light, it emits visible light at a specific wavelength, producing different colors. Fluorophores may be added as a molecular tag to different portions of a cell. DAPI is a fluorophore that specifically binds to DNA and fluoresces blue. In addition, immunofluorescence may be used as a laboratory technique to tag cells with specific fluorophores using antibodies, immune proteins created by B lymphocytes. Antibodies are utilized by the immune system in the identification and binding of foreign substances. Tubulin is a monomer of microtubules that compose the cellular cytoskeleton. The antibody anti-tubulin specifically binds to these tubulin monomeric subunits. A fluorophore can be chemically attached to the anti-tubulin antibody, which then fluoresces green. Numerous antibodies may bind to microtubules in order to amplify the fluorescent signal. Fluorescence microscopy allows for the observation of different components of the cell against a dark background for high intensity and specificity.
Practical Applications
Detection
Chromatin bridges are easiest and most readily visible when observing chromosomes stained with DAPI. DNA bridges appear to be a blue, “string-like” connection between two separated daughter cells. This effect is created when sticky ends of chromosomes remain connected to one another, even after mitosis. A chromatin bridge may also be observed using indirect immunofluorescence, in which anti-tubulin emits a green coloration when bound to microtubules in the presence of UV light. Because microtubules maintain the positions of the chromosomes during mitosis, they appear to be densely pinched between the two dividing, daughter cells. Chromatin bridges can be difficult to locate utilizing fluorescence microscopy, as this phenomenon is not incredibly abundant and tend to appear faint against the dark background.
Cancer
Recently, chromatin bridges have been implied as a diagnostic marker for cancer, while having been linked to tumorigenesis in humans.[8] This premise is based on the fact that as the mitotic cell divides and the daughter cells move further apart, stress on the DNA bridge leads to breakages in the chromosome at random points. As previously stated, the disruptions in the chromosome may lead to single chromosome mutations, including deletion, duplication and inversion, among others. This instability, defined as frequent changes in chromosomal structure and number, may be the basis of the development of cancer. While the frequency of chromatin bridges may be greater in tumor cells relative to normal cells, it may not be practical to utilize this phenomenon as a diagnostic tool. The process of staining and mounting sample cells using indirect immunofluorescence is time consuming. Even though DAPI staining is quick, neither laboratory technique can guarantee the presence of the bridges under the fluorescence microscope. The rarity of chromatin bridges, even in cancerous cells, makes this phenomenon difficult to be widely accepted diagnostic marker for cancer.
References
- ^ Chan KL, Hickson ID (2011). "New insights into the formation and resolution of ultra-fine anaphase bridges". Semin Cell Dev Biol.. http://www.sciencedirect.com/science/article/pii/S1084952111000875.
- ^ Hoffelder D, Luo L, Burke N, Watkins S, Gollin S, Saunders W (2004). "Resolution of anaphase bridges in cancer cells". Chromosoma 112. http://www.pitt.edu/~biohome/Dept/pdf/1621.pdf.
- ^ Gisselsson D, Jonson T, Yu C, Martins C, Mandahl N, Weigant J, Jin Y, Mertens F, Jin C (2002). "Centrosomal abnormalities, multipolar mitoses, and chromosomal instability in head and neck tumours with dysfunctional telomeres". British Journal of Cancer 87. http://www.nature.com/bjc/journal/v87/n2/full/6600438a.html.
- ^ Tkemaladze J, Chinchinadze K (2010). "Centriole, differentiation, and senescence". Rejuvenation Res 13 (2-3). http://www.liebertonline.com/doi/pdf/10.1089/rej.2009.0904.
- ^ de Lange T (2002). "Protection of mammalian telomeres". Oncogene 21 (4). http://www.nature.com/onc/journal/v21/n4/full/1205080a.html.
- ^ Genesca A, Pampalona J, Frias C, Dominguez d, Tusell L (2011). "Role of telomere dysfunction in genetic intratumor diversity". Adv Cancer Res 112. http://www.researchgate.net/publication/51649729_Role_of_telomere_dysfunction_in_genetic_intratumor_diversity.
- ^ Saunders W, Shuster M, Huang X, Gharaibeh B, Enyenihi A, Petersen I, Gollin S (1999). "Chromosomal instability and cytoskeletal defects in oral cancer cells". PNAS 97 (1). http://www.pnas.org/content/97/1/303.full.pdf+html.
- ^ Jallepalli PV, Lengauer C (2001). "Chromosome segregation and cancer: cutting through the mystery.". Nat. Rev Cancer 1 (2). http://www.nature.com/nrc/journal/v1/n2/full/nrc1101-109a.html.
Categories:- Chromosomes
- Cell biology
- Mitosis
Wikimedia Foundation. 2010.
