- Z-DNA
-
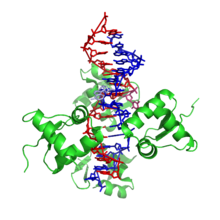
 The Z-DNA structure.Proteopedia Z-DNA
The Z-DNA structure.Proteopedia Z-DNAZ-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the double helix winds to the left in a zig-zag pattern (instead of to the right, like the more common B-DNA form). Z-DNA is thought to be one of three biologically active double helical structures along with A- and B-DNA.
Contents
History
Z-DNA was the first single-crystal X-ray structure of a DNA fragment (a self-complementary DNA hexamer d(CG)3). It was resolved as a left-handed double helix with two anti-parallel chains that were held together by Watson-Crick base pairs (see: x-ray crystallography). It was solved by Andrew Wang, Alexander Rich, and co-workers in 1979 at MIT.[1] The crystallisation of a B- to Z-DNA junction in 2005[2] provided a better understanding of the potential role Z-DNA plays in cells. Whenever a segment of Z-DNA forms, there must be B-Z junctions at its two ends, interfacing it to the B-form of DNA found in the rest of the genome.
In 2007, the RNA version of Z-DNA, Z-RNA, was described as a transformed version of an A-RNA double helix into a left-handed helix.[3] The transition from A-RNA to Z-RNA, however, was already described in 1984.[4]
Structure
Z-DNA is quite different from the right-handed forms. In fact, Z-DNA is often compared against B-DNA in order to illustrate the major differences. The Z-DNA helix is left-handed and has a structure that repeats every 2 base pairs. The major and minor grooves, unlike A- and B-DNA, show little difference in width. Formation of this structure is generally unfavourable, although certain conditions can promote it; such as alternating purine-pyrimidine sequence (especially poly(dGC)2), negative DNA supercoiling or high salt and some cations (all at physiological temperature, 37°C, and pH 7.3-7.4). Z-DNA can form a junction with B-DNA (called a "B-to-Z junction box") in a structure which involves the extrusion of a base pair.[5] The Z-DNA conformation has been difficult to study because it does not exist as a stable feature of the double helix. Instead, it is a transient structure that is occasionally induced by biological activity and then quickly disappears.[6]
Predicting Z-DNA structure
It is possible to predict the likelihood of a DNA sequence forming a Z-DNA structure. An algorithm for predicting the propensity of DNA to flip from the B-form to the Z-form, ZHunt, was written by Dr. P. Shing Ho in 1984 (at MIT).[7] This algorithm was later developed by Tracy Camp, P. Christoph Champ, Sandor Maurice, and Jeffrey M. Vargason for genome-wide mapping of Z-DNA (with P. Shing Ho as the principal investigator).[8]
Z-Hunt is available at Z-Hunt online.
Biological significance
While no definitive biological significance of Z-DNA has been found, it is commonly believed to provide torsional strain relief (supercoiling) while DNA transcription occurs.[2][9] The potential to form a Z-DNA structure also correlates with regions of active transcription. A comparison of regions with a high sequence-dependent, predicted propensity to form Z-DNA in human chromosome 22 with a selected set of known gene transcription sites suggests there is a correlation.[8]
Z-DNA formed after transcription initiation in some cases may be bound by RNA modifying enzymes, such as ADAR1[citation needed], which then alter the sequence of the newly formed RNA.[10]
In 2003, Biophysicist Alexander Rich of the Massachusetts Institute of Technology noticed that a poxvirus virulence factor, called E3L, mimicked a mammalian protein that binds Z-DNA.[11][12] In 2005, Rich and his colleagues pinned down what E3L does for the poxvirus. When expressed in human cells, E3L increases by five- to 10-fold the production of several genes that block a cell’s ability to self-destruct in response to infection.
Rich speculates that the Z-DNA is necessary for transcription and that E3L stabilizes the Z-DNA, thus prolonging expression of the anti-apoptotic genes. He suggests that a small molecule that interferes with the E3L binding to Z-DNA could thwart the activation of these genes and help protect people from pox infections.
Comparison Geometries of Some DNA Forms
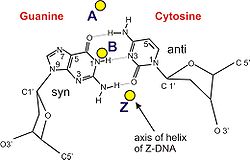
Geometry attribute A-form B-form Z-form Helix sense right-handed right-handed left-handed Repeating unit 1 bp 1 bp 2 bp Rotation/bp 32.7° 35.9° 60°/2 bp/turn 11 10.5 12 Inclination of bp to axis +19° −1.2° −9° Rise/bp along axis 2.3 Å (0.23 nm) 3.32 Å (0.332 nm) 3.8 Å (0.38 nm) Pitch/turn of helix 28.2 Å (2.82 nm) 33.2 Å (3.32 nm) 45.6 Å (4.56 nm) Mean propeller twist +18° +16° 0° Glycosyl angle anti anti C: anti,
G: synSugar pucker C3'-endo C2'-endo C: C2'-endo,
G: C3'-endoDiameter 23 Å (2.3 nm) 20 Å (2.0 nm) 18 Å (1.8 nm) Sources:[13][14][15] See also
- Mechanical properties of DNA
- DNA supercoil
- DNA
- A-DNA
- B-DNA
- Z-DNA binding protein 1 (ZBP1)
- Zuotin
- E3L
- ADAR1
- Proteopedia Z-DNA
References
- ^ Wang AHJ, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, Van der Marel G, Rich A (1979). "Molecular structure of a left-handed double helical DNA fragment at atomic resolution". Nature (London) 282 (5740): 680–686. Bibcode 1979Natur.282..680W. doi:10.1038/282680a0. PMID 514347.
- ^ a b Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK (2005). "Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases". Nature 437 (7062): 1183–1186. Bibcode 2005Natur.437.1183H. doi:10.1038/nature04088. PMID 16237447.
- ^ Placido D, Brown BA 2nd, Lowenhaupt K, Rich A, Athanasiadis A (2007). "A left-handed RNA double helix bound by the Zalpha domain of the RNA-editing enzyme ADAR1". Structure 15 (4): 395–404. doi:10.1016/j.str.2007.03.001. PMC 2082211. PMID 17437712. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2082211.
- ^ Hall K, Cruz P, Tinoco I Jr, Jovin TM, van de Sande JH (October 1984). "'Z-RNA'--a left-handed RNA double helix". Nature 311 (5986): 584–586. doi:10.1038/311584a0. PMID 6482970.
- ^ de Rosa M, de Sanctis D, Rosario AL, Archer M, Rich A, Athanasiadis A, Carrondo MA (2010-05-18). "Crystal structure of a junction between two Z-DNA helices". Proc Natl Acad Sci USA 107 (20): 9088–9092. doi:10.1073/pnas.1003182107. PMC 2889044. PMID 20439751. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2889044.
- ^ Zhang H, Yu H, Ren J, Qu X (2006). "Reversible B/Z-DNA transition under the low salt condition and non-B-form polydApolydT selectivity by a cubane-like europium-L-aspartic acid complex". Biophysical Journal 90 (9): 3203–3207. doi:10.1529/biophysj.105.078402. PMC 1432110. PMID 16473901. http://www.biophysj.org/cgi/content/full/90/9/3203.
- ^ Ho PS, Ellison MJ, Quigley GJ, Rich A (1986). "A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences". EMBO Journal 5 (10): 2737–2744. PMC 1167176. PMID 3780676. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1167176.
- ^ a b Champ PC, Maurice S, Vargason JM, Camp T, Ho PS (2004). "Distributions of Z-DNA and nuclear factor I in human chromosome 22: a model for coupled transcriptional regulation". Nucleic Acids Res. 32 (22): 6501–6510. doi:10.1093/nar/gkh988. PMC 545456. PMID 15598822. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=15598822.
- ^ Rich A, Zhang S (2003). "Timeline: Z-DNA: the long road to biological function". Nature Review Genetics 4 (7): 566–572. doi:10.1038/nrg1115. PMID 12838348.
- ^ Halber D (1999-09-11). "Scientists observe biological activities of 'left-handed' DNA". MIT News Office. http://web.mit.edu/newsoffice/1999/zdna-0911.html. Retrieved 2008-09-29.
- ^ Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A (2003). "A role for Z-DNA binding in vaccinia virus pathogenesis". Proc Natl Acad Sci USA 100 (12): 6974–6979. doi:10.1073/pnas.0431131100. PMC 165815. PMID 12777633. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=165815.
- ^ Kim YG, Lowenhaupt K, Oh DB, Kim KK, Rich A (2004). "Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: Implications for development of a therapy for poxvirus infection". Proc Natl Acad Sci USA 101 (6): 1514–1518. doi:10.1073/pnas.0308260100. PMC 341766. PMID 14757814. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=341766.
- ^ Sinden, Richard R (1994-01-15). DNA structure and function (1st ed.). Academic Press. pp. 398. ISBN 0-12-645750-6.
- ^ Rich A, Norheim A, Wang AHJ (1984). "The chemistry and biology of left-handed Z-DNA". Annual Review of Biochemistry 53 (1): 791–846. doi:10.1146/annurev.bi.53.070184.004043. PMID 6383204.
- ^ Ho PS (1994-09-27). "The non-B-DNA structure of d(CA/TG)n does not differ from that of Z-DNA". Proc Natl Acad Sci USA 91 (20): 9549–9553. doi:10.1073/pnas.91.20.9549. PMC 44850. PMID 7937803. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=44850.
External links
Types of nucleic acids Constituents Nucleobases · Nucleosides · Nucleotides · DeoxynucleotidesRibonucleic acids
(coding and non-coding)translation: mRNA (pre-mRNA/hnRNA) · tRNA · rRNA · tmRNA
regulatory: miRNA · siRNA · piRNA · aRNA · RNAi ·
RNA processing: snRNA · snoRNA
other/ungrouped: gRNA · shRNA · stRNA · ta-siRNADeoxyribonucleic acids Nucleic acid analogues Cloning vectors biochemical families: prot · nucl · carb (glpr, alco, glys) · lipd (fata/i, phld, strd, gllp, eico) · amac/i · ncbs/i · ttpy/iCategories:
Wikimedia Foundation. 2010.