- DNA-encoded chemical library
-
DNA-encoded chemical libraries (DEL) are a new technology for the synthesis and screening of collections of chemical compounds of unprecedented size and quality. DEL represents an advance in medicinal chemistry which bridges the fields of combinatorial chemistry and molecular biology. The driving force for the development of DEL technology is to improve and streamline the drug discovery process in particular early phase discovery activities such as target validation and hit discovery.
DEL technology involves the conjugation of chemical compounds or building blocks to short DNA fragments that serve as identification bar codes and in some cases also direct and control the chemical synthesis. The technique, in principle, enables the mass creation and interrogation of libraries via affinity selection, typically on an immobilized protein target. In contrast to conventional screening procedures such as high-throughput screening, biochemical assays are not required for binder identification, in principle allowing the isolation of binders to a wide range of proteins historically difficult to tackle with conventional screening technologies. So, in addition to the general discovery of target specific molecular compounds, the availability of binders to pharmacologically important, but so-far “undruggable” target proteins opens new possibilities to develop novel drugs for diseases that could not be treated so far. In eliminating the requirement to initially assess the activity of hits it is hoped and expected that many of the high affinity binders identified will be shown to be active in independent analysis of selected hits, therefore offering an efficient method to identify high quality hits and pharmaceutical leads.
Contents
DNA-encoded chemical libraries and display technologies
Until recently, the application of molecular evolution in the laboratory had been limited to display technologies involving biological molecules, where small molecules lead discovery was considered beyond this biological approach. DEL has opened the field of display technology to include non-natural compounds such as small molecules, extending the application of molecular evolution and natural selection to the identification of small molecule compounds of desired activity and function. DNA encoded chemical libraries bear resemblance to biological display technologies such as antibody phage display technology, yeast display, mRNA display and aptamer SELEX. In antibody phage display, antibodies are physically linked to phage particles that bear the gene coding for the attached antibody, which is equivalent to a physical linkage of a “phenotype” (the protein) and a “genotype” (the gene encoding for the protein ).[1] Phage-displayed antibodies can be isolated from large antibody libraries by mimicking molecular evolution: through rounds of selection (on an immobilized protein target), amplification and translation.[2] In DEL the linkage of a small molecule to an identifier DNA code allows the facile identification of binding molecules. DEL libraries are subjected to affinity selection procedures on an immobilized target protein of choice, after which non-binders are removed by washing steps, and binders can subsequently be amplified by polymerase chain reaction (PCR) and identified by virtue of their DNA code (e.g.by DNA sequencing). In evolution-based DEL technologies (see below) hits can be further enriched by performing rounds of selection, PCR amplification and translation in analogy to biological display systems such as antibody phage display. This makes it possible to work with much larger libraries.
History
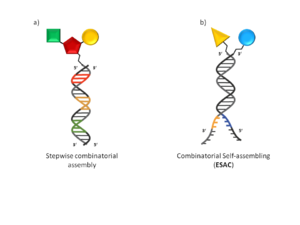
Fig. 1 DNA-encoded library displaying chemical compounds Schematic representation of DNA-encoded library displaying chemical compounds directly attached to oligonucleotides. a) Library generated by “stepwise combinatorial” assembling presenting a single oligonucleotide covalently linked to a putative binding molecule. b) Library construct in “combinatorial self-assembling” fashion (Encoded Self-Assembling Chemical library). Multiple pairing oligonucleotides display a covalently linked binding molecule
The concept of DNA-encoding was first described in a theoretical paper by Brenner and Lerner in 1992 in which was proposed to link each molecule of a chemically synthesized entity to a particular oligonucleotide sequence constructed in parallel and to use this encoding genetic tag to identify and enrich active compounds.[3] In 1993 the first practical implementation of this approach was presented by S. Brenner and K. Janda and similarly by the group of M.A. Gallop.[4][5] Brenner and Janda suggested to generate individual encoded library members by an alternating parallel combinatorial synthesis of the heteropolymeric chemical compound and the appropriate oligonucleotide sequence on the same bead in a “split-&-pool”-based fashion (see below).[4]
Since unprotected DNA is restricted to a narrow window of conventional reaction conditions, until the end of 1990s a number of alternative encoding strategies were envisaged (i.e. MS-based compound tagging, peptide encoding, haloaromatic tagging, encoding by secondary amines, semiconductor devices.), mainly to avoid inconvenient solid phase DNA synthesis and to create easily screenable combinatorial libraries in high-throughput fashion.[6] However, the selective amplifiability of DNA greatly facilitates library screening and it becomes indispensable for the encoding of organic compounds libraries of this unprecedented size. Consequently, at the beginning of 2000s DNA-combinatorial chemistry experienced a revival.
The beginning of the millennium saw the introduction of several independent developments in DEL technology. These technologies can be classified under two general categories: non-evolution-based and evolution-based DEL technologies capable of molecular evolution. The first category benefits from the ability to use off the shelf reagents and therefore enables rather straighforward library generation. Hits can be identified by DNA sequencing, however DNA translation and therefore molecular evolution is not feasible by these methods. The split and pool approaches developed by researchers at Praecis Pharmaceuticals (now owned by GlaxoSmithKline), Nuevolution (Copenhagen, Denmark) and ESAC technology developed in the laboratory of Prof D. Neri (Institute of Pharmaceutical Science, Zurich, Switzerland) fall under this category. ESAC technology sets itself apart being a combinatorial self-assembling approach which resembles fragment based hit discovery (Fig 1b). Here DNA annealing enables discrete building block combinations to be sampled, but no chemical reaction takes place between them. Examples of evolution-based DEL technologies are DNA-routing developed by Prof. D.R. Halpin and Prof. P.B. Harbury (Stanford University, Stanford, CA), DNA-templated synthesis developed by Prof. D. Liu (Harvard University, Cambridge, MA) and commercialized by Ensemble Discovery (Cambridge, MA) and YoctoReactor technology[7] developed and commercialized by Vipergen (Copenhagen, Denmark). These technologies are described in further detail below. DNA-templated synthesis and YoctoReactor technology require the prior conjugation of chemical building blocks (BB) to a DNA oligonucleotide tag before library assembly, therefore more upfront work is required before library assembly. Furthermore, the DNA tagged BBs enable the generation of a genetic code for synthesized compounds and artificial translation of the genetic code is possible: That is the BB’s can be recalled by the PCR-amplified genetic code, and the library compounds can be regenerated. This, in turn, enables the principle of Darwinian natural selection and evolution to be applied to small molecule selection in direct analogy to biological display systems; through rounds of selection, amplification and translation.
Non-evolution based technologies
Split-&-Pool DNA Encoding
In order to apply combinatorial chemistry for the synthesis of DNA-encoded chemical libraries, a Split-&-Pool approach was pursued. Initially a set of unique DNA-oligonucleotides (n) each containing a specific coding sequence is chemically conjugated to a corresponding set of small organic molecules.Consequently the oligonucleotide-conjugate compounds are mixed ("Pool") and divided("Split")into a number of groups (m). In appropriate conditions a second set of building blocks (m) are coupled to the first one and a further oligonucleotide which is coding for the second modification is enzymatically introduced before mixing again. This “split-&-pool” steps can be iterated a number of times (r) increasing at each round the library size in a combinatorial manner (i.e. (n x m)r).
Stepwise coupling of coding DNA fragments to nascent organic molecules
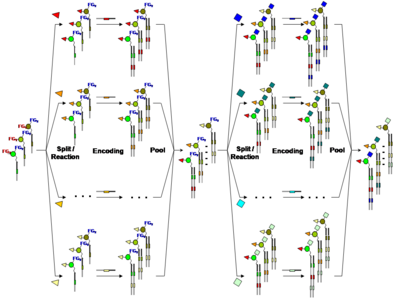 Fig. 3 DNA-encoded library by "Split-&-Pool stepwise coupling of coding DNA fragments to nascent organic molecules An initial set of multifunctional building blocks (FGn represents the different orthogonal functional groups) are covalently conjugated to a corresponding encoding oligonucleotide and reacted in a split-&-pool fashion on a specific functional group (FG1 in red) with a suitable collection of reagents. Following enzymatic encoding, a further round of split-&-pool is initiated. At this stage the second functional group (FG2 in blue) undergoes an additional reaction step with a different set of suitable reagents. The identity of the final modification could be ensured yet again by enzymatic DNA encoding by means of a further oligonucleotide carrying a specific coding region.
Fig. 3 DNA-encoded library by "Split-&-Pool stepwise coupling of coding DNA fragments to nascent organic molecules An initial set of multifunctional building blocks (FGn represents the different orthogonal functional groups) are covalently conjugated to a corresponding encoding oligonucleotide and reacted in a split-&-pool fashion on a specific functional group (FG1 in red) with a suitable collection of reagents. Following enzymatic encoding, a further round of split-&-pool is initiated. At this stage the second functional group (FG2 in blue) undergoes an additional reaction step with a different set of suitable reagents. The identity of the final modification could be ensured yet again by enzymatic DNA encoding by means of a further oligonucleotide carrying a specific coding region.
A promising strategy for the construction of DNA-encoded libraries is represented by the use of multifunctional building blocks covalently conjugate to an oligonucleotide serving as a “core structure” for library synthesis. In a ‘pool-and-split’ fashion a set of multifunctional scaffolds undergo orthogonal reactions with series of suitable reactive partners. Following each reaction step, the identity of the modification is encoded by an enzymatic addition of DNA segment to the original DNA “core structure”.[8][9] The use of N-protected amino acids covalently attached to a DNA fragment allow, after a suitable deprotection step, a further amide bond formation with a series of carboxylic acids or a reductive amination with aldehydes. Similarly, diene carboxylic acids used as scaffolds for library construction at the 5’-end of amino modified oligonucleotide, could be subjected to a Diels-Alder reaction with a variety of maleimide derivatives. After completion of the desired reaction step, the identity of the chemical moiety added to the oligonucleotide is established by the annealing of a partially complementary oligonucleotide and by a subsequent Klenow fill-in DNA-polymerization, yielding a double stranded DNA fragment. The synthetic and encoding strategies described above enable the facile construction of DNA-encoded libraries of a size up to 104 member compounds carrying two sets of “building blocks”. However the stepwise addition of at least three independent sets of chemical moieties to a tri-functional core building block for the construction and encoding of a very large DNA-encoded library (comprising up to 106 compounds) can also be envisaged.[8](Fig.2)
Combinatorial self-assembling
Encoded Self-Assembling Chemical libraries
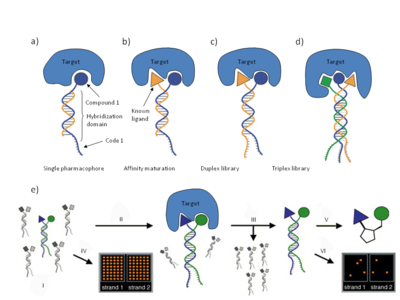 Fig. 4 ESAC library technology overview Small organic molecules are coupled to 5’-amino modified oligonucleotides, containing a hybridization domain and a unique coding sequence, which ensure the identity of the coupled molecule. The ESAC library can be used in single pharmacophore format (a), in affinity maturations of known binders (b), or in de novo selections of binding molecules by self assembling of sublibraries in DNA-double strand format (c) as well as in DNA-triplexes (d). The ESAC library in the selected format is used in a selection and read-out procedure (e). Following incubation of the library (i) with the target protein of choice (ii) and washing of unbound molecules (iii), the oligonucleotide codes of the binding compounds are PCR-amplified and compared with the library without selection on oligonucleotide micro-arrays (iv, v). Identified binders/binding pairs are validated after conjugation (if appropriate) to suitable scaffolds (vi).
Fig. 4 ESAC library technology overview Small organic molecules are coupled to 5’-amino modified oligonucleotides, containing a hybridization domain and a unique coding sequence, which ensure the identity of the coupled molecule. The ESAC library can be used in single pharmacophore format (a), in affinity maturations of known binders (b), or in de novo selections of binding molecules by self assembling of sublibraries in DNA-double strand format (c) as well as in DNA-triplexes (d). The ESAC library in the selected format is used in a selection and read-out procedure (e). Following incubation of the library (i) with the target protein of choice (ii) and washing of unbound molecules (iii), the oligonucleotide codes of the binding compounds are PCR-amplified and compared with the library without selection on oligonucleotide micro-arrays (iv, v). Identified binders/binding pairs are validated after conjugation (if appropriate) to suitable scaffolds (vi).
Encoded Self-Assembling Chemical (ESAC) libraries rely on the principle that two sublibraries of a size of x members (e.g. 103) containing a constant complementary hybridization domain can yield a combinatorial DNA-duplex library after hybridization with a complexity of x2 uniformly represented library members (e.g. 106).[10] Each sub-library member would consist of an oligonucleotide containing a variable, coding region flanked by a constant DNA sequence, carrying a suitable chemical modification at the oligonucleotide extremity.[10] The ESAC sublibraries can be used in at least four different embodiments.[10]
- A sub-library can be paired with a complementary oligonucleotide and used as a DNA encoded library displaying a single covalently linked compound for affinity-based selection experiments.
- A sub-library can be paired with an oligonucleotide displaying a known binder to the target, thus enabling affinity maturation strategies.
- Two individual sublibraries can be assembled combinatorially and used for the de novo identification of bindentate binding molecules.
- Three different sublibraries can be assembled to form a combinatorial triplex library.
Preferential binders isolated from an affinity-based selection can be PCR-amplified and decoded on complementary oligonucleotide microarrays[11] or by concatenation of the codes, subcloning and sequencing.[12] The individual building blocks can eventually be conjugated using suitable linkers to yield a drug-like high-affinity compound. The characteristics of the linker (e.g. length, flexibility, geometry, chemical nature and solubility) influence the binding affinity and the chemical properties of the resulting binder.(Fig.3)
Bio-panning experiments on HSA of a 600-member ESAC library allowed the isolation of the 4-(p-iodophenyl)butanoic moiety. The compound represents the core structure of a series of portable albumin binding molecules and of AlbufluorTM a recently developed fluorescein angiographic contrast agent currently under clinical evaluation.[13]
ESAC technology has been used for the isolation of potent inhibitors of bovine trypsin and for the identification of novel inhibitors of stromelysin-1 (MMP-3) , a matrix metalloproteinase involved in both physiological and pathological tissue remodeling processes, as well as in disease processes, such as arthritis and metastasis.[14][15]
Evolution-based technologies
DNA-routing
In 2004, D.R. Halpin and P.B. Harbury presented a novel intriguing method for the construction of DNA-encoded libraries. For the first time the DNA-conjugated templates served for both encoding and programming the infrastructure of the “split-&-pool” synthesis of the library components.[16] The design of Halpin and Harbury enabled alternating rounds of selection, PCR amplification and diversification with small organic molecules, in complete analogy to phage display technology. The DNA-routing machinery consists of a series of connected columns bearing resin-bound anticodons, which could sequence-specifically separate a population of DNA-templates into spatially distinct locations by hybridization.[16] According to this split-and-pool protocol a peptide combinatorial library DNA-encoded of 106 members was generated.[17]
DNA-templated synthesis
Further information: Nucleic acid templated chemistry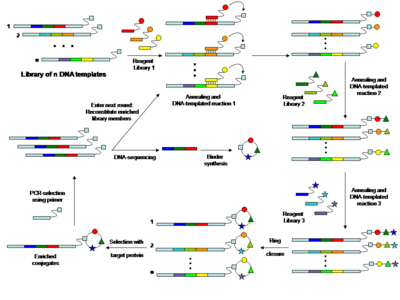 Fig. 2 DNA-encoded library by ‘DNA-templated synthesis’A library of oligonucleotides (i.e. 64 different oligonucleotides) containing three coding regions was hybridized to a library of reagent compound-oligonucleotide conjugates (i.e. 4 reagent oligonucleotide conjugates), able of pairing with the initial coding domain of the template oligonucleotide. After transferring of the compounds on the corresponding olgonucleotide template, the synthesis cycle was repeated the desired number of times with further sets of carrier compound-oligonucleotide conjugates (i.e. two rounds with four carrier compound-oligonucleotide conjugates per round). Subsequently functional selection was performed and the sequence of the binding template amplified by PCR. Thus, DNA-sequencing allowed the identification of the binding molecule.
Fig. 2 DNA-encoded library by ‘DNA-templated synthesis’A library of oligonucleotides (i.e. 64 different oligonucleotides) containing three coding regions was hybridized to a library of reagent compound-oligonucleotide conjugates (i.e. 4 reagent oligonucleotide conjugates), able of pairing with the initial coding domain of the template oligonucleotide. After transferring of the compounds on the corresponding olgonucleotide template, the synthesis cycle was repeated the desired number of times with further sets of carrier compound-oligonucleotide conjugates (i.e. two rounds with four carrier compound-oligonucleotide conjugates per round). Subsequently functional selection was performed and the sequence of the binding template amplified by PCR. Thus, DNA-sequencing allowed the identification of the binding molecule.
In 2001 David Liu and co-workers showed that complementary DNA oligonucleotides can be used to assist certain synthetic reactions, which do not efficiently take place in solution at low concentration.[18][19] A DNA-heteroduplex was used to accelerate the reaction between chemical moieties displayed at the extremities of the two DNA strands. Furthermore, the "proximity effect", which accelerates bimolecular reaction, was shown to be distance-independent (at least within a distance of 30 nucleotides).[18][19] In a sequence-programmed fashion oligonucleotides carrying one chemical reactant group were hybridized to complementary oligonucleotide derivatives carrying a different reactive chemical group. The proximity conferred by the DNA hybridization drastically increases the effective molarity of the reaction reagents attached to the oligonucleotides, enabling the desired reaction to occur even in an aqueous environment at concentrations which are several orders of magnitude lower than those needed for the corresponding conventional organic reaction not DNA-templated.[20] Using a DNA-templated set-up and sequence-programmed synthesis Liu and co-workers generated a 64 member compound DNA encoded library of macrocycles.[21]
3-Dimensional proximity-based technology (YoctoReactor technology)
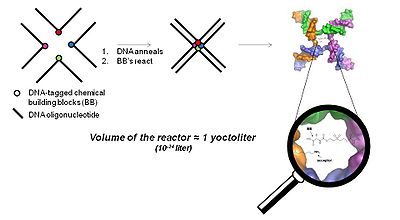
The YoctoReactor (yR) is a 3D proximity-driven approach which exploits the self-assembling nature of DNA oligonucleotides into 3, 4 or 5-way junctions to direct small molecule synthesis at the center of the junction. Figure 5 illustrates the basic concept with a 4-way DNA junction.
Fig. 5 Fundamental principle of the YoctoReactor. The center of 3, 4 and 5 way DNA junctions (a 4-way junction is shown here) becomes a yoctoliter-scale reactor where small molecule synthesis is facilitated in what has been termed the YoctoReactor (yR). Colored circles depict the chemical building blocks (BB) which are attached to carefully designed DNA oligonucleotides (black lines). Upon DNA annealing the BB are brought into proximity at the center of the DNA junction where they undergo chemical reaction.
The center of the DNA junction constitutes a volume on the order of a yoctoliter, hence the name YoctoReactor. This volume contains a single molecule reaction yielding reaction concentrations in the high mM range. The effective concentration facilitated by the DNA greatly accelerates chemical reactions that otherwise would not take place at the actual concentration several orders of magnitude lower.
Building a yR library
Figure 6 illustrates the generation of a yR library using a 3-way DNA junction.
Fig. 6 YoctoReactor library assembly. Stepwise assembly of a DEL library using YoctoReactor technology. A 3-way reactor is shown here. (a) Position 1 (P1) and P2 BB are brought into proximity and undergo a chemical reaction in the presence of a helper oligonucleotide in P3. (b) The structure is purified by polyacrylamide gel electrophoresis (PAGE), the P1 and P2 DNA is ligated and the P2 linker is cleaved. (c) P3 BB is annealed to the P1-P2 ligation product from step b, and a chemical reaction between P2 and P3 BBs takes place. (d) The reaction product is purified by PAGE, the DNA is ligated and P3 linker is cleaved yielding a compound (OOO) covalently attached to the folded yR. (e) The yR is dismantled by primer extension yielding a double-stranded disply product exposing the reaction product for selection and molecular evolution.
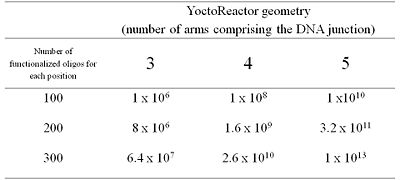
In summary, chemical building-blocks (BB) are attached via cleavable or non-cleavable linkers to three types of bispecific DNA oligonucleotides (oligo-BBs) representing each arm of the yR. To facilitate synthesis in a combinatorial manner, the oligo-BBs are designed such that the DNA contains (a) the code for an attached BB at the distal end of the oligo (colored lines) and (b) areas of constant DNA sequence (black lines) to bring about the self assembly of the DNA into a 3-way junction (independently of the BB) and the subsequent chemical reaction. Chemical reactions are performed via a stepwise procedure and after each step the DNA is ligated and the product purified by polyacryamide gel electrophoresis. Cleavable linkers (BB-DNA) are used for all but one position yielding a library of small molecules with a single covalent link to the DNA code. Table 1 outlines how libraries of different sizes can be generated using yR technology.
The yR design approach provides an unvarying reaction site with regard to both (a) distance between reactants and (b) sequence environment surrounding the reaction site. Furthermore the intimate connection between the code and the BB on the oligo-BB moieties which are mixed combinatorially in a single pot confers a high fidelity to the encoding of the library. The code of the synthesized products, furthermore, is not preset, but rather is assembled combinatorially and synthesized in synchronicity with the innate product.
Rolling Translation
The latter two features of yR technology described above are important in that they enable a translational process analogous to biological translation. The ability to translate is essential to yR technology and the manipulation of very large libraries, however biological translation is limited to biological molecules; the translation of DNA into protein. With the yR an artificial translation system, termed Rolling Translation, has been established: The information for the synthesis of the small molecule is contained in the attached DNA (genotype) and the molecule-genetic code (phenotype-genotype) can be regenerated via Rolling Translation.[22]
Molecular evolution for hit identification
Using principles of Darwinian selection, high affinity hits are identified from the yR library via affinity selection on an immobilized target in rounds of selection, PCR amplification and translation. Affinity selection, described in detail elsewhere[ref], consists of incubating the immobilized target (typically a protein immobilized via a tag to a magnetic bead) with the library, during which time library compounds have the opportunity to bind to the target. Subsequent washing steps remove non-binders, reduce the level of unspecific binders and remove the vast majority of the library members. Elution of the mixture of non-binders and non-specific binders under denaturing conditions yields a mixture, which ideally is now enriched for binders to the target of interest. The mixture is subjected to PCR amplification followed by artificial translation after which a second round of selection can be performed. Typically if very large libraries are being interrogated (106-109), 2-3 rounds of selection are required to enrich binders above background. Workers at Vipergen (Copenhagen, Denmark), the company commercializing the yR, have reported a selective enrichment of a known peptide binder to anti-Enkephalin antibody from a frequency of 1 in 100 million in a model library to a final frequency of 1.7% in only two rounds of selection.[22]The creation and manipulation of libraries up to 1x 1012 in size is therefore conceptually possible with the YoctoReactor technology.
Decoding of DNA-encoded chemical libraries
Following selection from DNA-encoded chemical libraries, the decoding strategy for the fast and efficient identification of the specific binding compounds is crucial for the further development of the DEL technology. So far, Sanger-sequencing-based decoding, microarray-based methodology and high-throughput sequencing techniques represented the main methodologies for the decoding of DNA-encoded library selections.
Sanger sequencing-based decoding
Although many authors implicitly envisaged a traditional Sanger sequencing-based decoding,[4][5][10][17][21] the number of codes to sequence simply according to the complexity of the library is definitely an unrealistic task for a traditional Sanger sequencing approach. Nevertheless, the implementation of Sanger sequencing for decoding DNA-encoded chemical libraries in high-throughput fashion was the first to be described.[10] After selection and PCR amplification of the DNA-tags of the library compounds, concatamers containing multiple coding sequences were generated and ligated into a vector. Following Sanger sequencing of a representative number of the resulting colonies revealed the frequencies of the codes present in the DNA-encoded library sample before and after selection.[10]
Microarray-based decoding
A DNA microarray is a device for high-throughput investigations widely used in molecular biology and in medicine. It consists of an arrayed series of microscopic spots (‘features’ or ‘locations’) containing few picomoles of oligonucleotides carrying a specific DNA sequence. This can be a short section of a gene or other DNA element that are used as probes to hybridize a DNA or RNA sample under suitable conditions. Probe-target hybridization is usually detected and quantified by fluorescence-based detection of fluorophore-labeled targets to determine relative abundance of the target nucleic acid sequences. Microarray has been used for the successfully decoding of ESAC DNA-encoded libraries.[10] The coding oligonucleotides representing the individual chemical compounds in the library, are spotted and chemically linked onto the microarray slides, using a BioChip Arrayer robot. Subsequently, the oligonucleotide tags of the binding compounds isolated from the selection are PCR amplified using a fluorescent primer and hybridized onto the DNA-microarray slide. Afterwards, microarrays are analyzed using a laser scan and spot intensities detected and quantified. The enrichment of the preferential binding compounds is revealed comparing the spots intensity of the DNA-microarray slide before and after selection.[10]
Decoding by high throughput sequencing
According to the complexity of the DNA encoded chemical library (typically between 103 and 106 members), a conventional Sanger sequencing based decoding is unlikely to be usable in practice, due both to the high cost per base for the sequencing and to the tedious procedure involved.[23] High throughput sequencing technologies exploited strategies that parallelize the sequencing process displacing the use of capillary electrophoresis and producing thousands or millions of sequences at once. In 2008 was described the first implementation of a high-throughput sequencing technique originally developed for genome sequencing (i.e. "454 technology") to the fast and efficient decoding of a DNA encoded chemical library comprising 4000 compounds.[8] This study led to the identification of novel chemical compounds with submicromolar dissociation constants towards streptavidin and definitely shown the feasibility to construct, perform selections and decode DNA-encoded libraries containing millions of chemical compounds.[8]
See also
References
- ^ G.P. Smith, Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface,Science 228 (1985), 1315–1317
- ^ H.R.Hoogenboom, Overview of antibody phage-display technology and its applications. Methods Mol.Biol. 178 (2002),1-37
- ^ S. Brenner and R.A. Lerner, Encoded combinatorial chemistry, Proc. Natl. Acad. Sci. U. S. A. 89 (1992), 5381–5383
- ^ a b c J. Nielsen, S. Brenner and K.D. Janda, Synthetic methods for the implementation of encoded combinatorial chemistry, J. Am. Chem. Soc. 115 (1993), 9812–9813
- ^ a b M.C. Needels, D.G. Jones, E.H. Tate, G.L. Heinkel, L.M. Kochersperger, W.J. Dower, R.W. Barrett and M.A. Gallop, Generation and screening of an oligonucleotide-encoded synthetic peptide library, Proc. Natl. Acad. Sci. U. S. A. 90 (1993), 10700–10704
- ^ Mukund S. Chorghade, Drug Discovery and Development - Combinatorial Chemistry in the Drug Discovery Process ISBN 978-0-471-39848-6 Ed. 2006 John Wiley & Sons, Inc., 129-167
- ^ T.R.Heitner and N.V.Hansen, Streamlining hit discovery and optimization with a yoctoliter scale DNA reactor. Exp. Opin. Drug. Disc. 4 (2009), 1201-1213
- ^ a b c d Mannocci L., Zhang Y., Scheuermann J., Leimbacher M., De Bellis G., Rizzi E., Dumelin C., Melkko S, Neri D., High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries, Proc. Natl. Acad. Sci. U. S. A. 105(46), (2008), 17670-17675
- ^ Buller F., Mannocci L., Zhang Y., Dumelin C.E., Scheuermann J., Neri D., Design and synthesis of a novel DNA-encoded chemical library using Diels-Alder cycloadditions, Bioorg Med. Chem. Lett. 18(22), (2008), 5926-5931
- ^ a b c d e f g h S. Melkko, J. Scheuermann, C.E. Dumelin and D. Neri, Encoded self-assembling chemical libraries, Nat. Biotechnol. 22 (2004), 568–574
- ^ M. Lovrinovic and C.M. Niemeyer, DNA microarrays as decoding tools in combinatorial chemistry and chemical biology, Angew Chem., Int. Ed. Engl. 44 (2005), 3179–3183
- ^ S. Melkko, J. Sobek, G. Guarda, J. Scheuermann, C.E. Dumelin and D. Neri, Encoded self-assembling chemical libraries, Chimia 59 (2005), 798–802
- ^ Dumelin CE, Trüssel S, Buller F, Trachsel E, Bootz F, Zhang Y, Mannocci L, Beck SC, Drumea-Mirancea M, Seeliger MW, Baltes C, Müggler T, Kranz F, Rudin M, Melkko S, Scheuermann J, Neri D. A portable albumin binder from a DNA-encoded chemical library. Angew Chem Int Ed Engl. 47(17) (2008); 3196-201
- ^ Melkko S, Zhang Y, Dumelin CE, Scheuermann J, Neri D., Isolation of high-affinity trypsin inhibitors from a DNA-encoded chemical library. Angew Chem Int Ed Engl. 46(25) (2007), 4671-4674
- ^ Scheuermann J, Dumelin CE, Melkko S, Zhang Y, Mannocci L, Jaggi M, Sobek J, Neri D., DNA-Encoded Chemical Libraries for the Discovery of MMP-3 Inhibitors. Bioconjug Chem. 19(3) (2008), 778-785
- ^ a b D.R. Halpin and P.B. Harbury, DNA display I. Sequence-encoded routing of DNA populations, PLoS Biol. 2 (2004), 1015–1021
- ^ a b D.R. Halpin and P.B. Harbury, DNA display. II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution, PLoS Biol. 2 (2004), 1022–1030
- ^ a b Z.J. Gartner and D.R. Liu, The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules, J. Am. Chem. Soc. 123 (2001), 6961–6963
- ^ a b C.T. Calderone, J.W. Puckett, Z.J. Gartner and D.R. Liu, Directing otherwise incompatible reactions in a single solution by using DNA-templated organic synthesis, Angew Chem., Int. Ed. Engl. 41 (2002), 4104–4108
- ^ X. Li and D.R. Liu, DNA-templated organic synthesis: nature's strategy for controlling chemical reactivity applied to synthetic molecules, Angew Chem., Int. Ed. Engl. 43 (2004), 4848–4870
- ^ a b Z.J. Gartner, B.N. Tse, R. Grubina, J.B. Doyon, T.M. Snyder and D.R. Liu, DNA-templated organic synthesis and selection of a library of macrocycles, Science 305 (2004), 1601–1605
- ^ a b M.H.Hansen, P.Blakskjær, et.al., A yoctoliter-scale DNA reactor for small-molecule evolution, J.Am.Chem.Soc. 131 (2009), 1322-1327
- ^ Sanger, F. , Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74 (1977), 5463–5467
Categories:- Biotechnology
- Scientific techniques
- Drug discovery
Wikimedia Foundation. 2010.