- Okazaki fragments
-
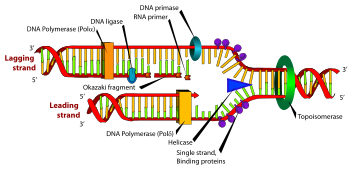
Okazaki fragments are short molecules of single-stranded DNA that are formed on the lagging strand during DNA replication. They are between 1,000 to 2,000 nucleotides long in Escherichia coli and are between 100 to 200 nucleotides long in eukaryotes.
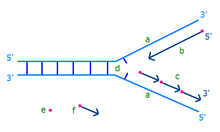
On the leading strand DNA replication proceeds continuously along the DNA molecule as the parent double-stranded DNA is unwound. But on the lagging strand the new DNA is made in installments, which are later joined together by a DNA ligase enzyme. This is because the enzymes that synthesise the new DNA can only work in one direction along the parent DNA molecule. On the leading strand this route is continuous, but on the lagging strand it is discontinuous.[1]
They were originally discovered in 1966 by Kiwako Sakabe and Reiji Okazaki in their research on DNA replication of Escherichia coli.[2] They were further investigated by them and their colleagues through their research including the study on bacteriophage DNA replication in Escherichia coli.[3][4][5]
Contents
Experiments
The work of Tsuneko and Reiji Okazaki provided experimental evidence supporting the hypothesis that DNA replication is a discontinuous process. Previously, it was commonly accepted that replication was continuous in both the 3’ to 5’ and 5’ to 3’ directions. 3’ and 5’ are specifically numbered carbons on the deoxyribose ring in nucleic acids, and refer to the orientation or directionality of a strand. In 1967, the Okazakis and their colleagues suggested that there is no found mechanism that showed continuous replication in the 3’ to 5’ direction, only 5’ to 3’ using DNA polymerase, a replication enzyme. The team hypothesized that if discontinuous replication was used, short strands of DNA, synthesized at the replicating point, could be attached in the 5’ to 3’ direction to the older strand.[5]
To distinguish the method of replication used by DNA experimentally, the team pulse-labeled newly replicated areas of Escherichia coli chromosomes, denatured, and extracted the DNA. A large amount of radioactive short units meant that the replication method was likely discontinuous. The hypothesis was further supported by the discovery of polynucleotide ligase, an enzyme that links short DNA strands together.[6]
In 1968, the Okazakis gathered additional evidence of nascent DNA strands. They hypothesized that if discontinuous replication, involving short DNA chains linked together by polynucleotide ligase, is the mechanism used in DNA synthesis, then “newly synthesized short DNA chains would accumulate in the cell under conditions where the function of ligase is temporarily impaired.” E.coli were infected with Bacteriophage T4 that produce temperature-sensitive polynucleotide ligase. The cells infected with the T4 Phages accumulated a large amount of short, newly synthesized DNA chains, as predicted in the hypothesis, when exposed to high temperatures. This experiment further supported the Okazakis’ hypothesis of discontinuous replication and linkage by polynucleotide ligase. It disproved the notion that short chains were produced during the extraction process as well.[7]
The Okazakis’ experiments provided extensive information on the replication process of DNA and the existence of short, newly synthesized DNA chains that later became known as Okazaki fragments.
Process
Pathways
There are two pathways that have been proposed to process Okazaki fragments. In the first pathway, only the nuclease FEN1, which cleaves the short flaps immediately when they form, is involved. While this pathway can process basically all flaps, an issue with this pathway is that some flaps may escape cleavage and thus become long. These flaps then bind to replication protein A (RPA) which inhibits FEN1 cleavage.[8] The second pathway thus becomes involved and is able to utilize both FEN1 and Dna2 nucleases to process the long flaps. Dna2 can cleave the RPA bound flap as it is able to displace to RPA, while creating a flat to which RPA cannot bind. Then, FEN1 will complete the cleavage of the flap. Dna2 is a key part of this process. Without the Dna2, the RPA bound flaps could not be processed which would ultimately lead to cell instability. The Pif1 helicase is also involved in this pathway as it aids creation of long flaps. Without the Pif1 helicase, the flaps would not become long enough to need cleavage by Dna2. Recently, it has been suggested that an alternative pathway for Okazaki fragment processing exists. This alternative pathway occurs when the Pif1 helicase removes entire Okazaki fragments initiated by fold back flaps.[9]
Alternate pathway
Until recently, there were only two known pathways to process Okazaki fragments. However, current investigations have concluded that a new pathway for Okazaki fragmentation and DNA replication exists. This alternate pathway involves the enzymes Pol δ with Pif1 which perform the same flap removal process as Polδ and FEN1.[9]
Enzymes involved in fragment formation
Primase
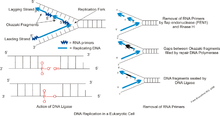
Main article: PrimasePrimase adds RNA primers onto the lagging strand, which allows synthesis of Okazaki fragments from 5’ to 3’. However, Primase creates RNA primers at a much lower rate than that at which DNA polymerase synthesizes DNA on the leading strand. DNA polymerase on the lagging strand also has to be continually recycled to construct Okazaki fragments following RNA primers. This makes the speed of lagging strand synthesis much lower than that of the leading strand. To solve this, primase acts as a temporary stop signal, briefly halting the progression of the replication fork during DNA replication. This molecular process prevents the leading strand from overtaking the lagging strand.[10]
DNA polymerase δ
Following creation of RNA primers by Primase on the lagging strand, DNA polymerase δ synthesizes Okazaki fragments. DNA polymerase δ also carries out a 3’ to 5’ exonuclease role, proofreading newly synthesized DNA strands during DNA replication. When the polymerase encounters an erroneous base pair, it removes one of the nucleotides and replaces it with a correct one. A third function of DNA polymerase δ is to supplement FEN1/RAD27 5’ Flap Endonuclease activity. This includes preventing and removing the strand displacement of 5’ flaps, and creating ligatable nicks at the border of Okazaki fragments.[11][12]
DNA ligase I
Main article: DNA ligaseDuring lagging strand synthesis, DNA ligase I connects the Okazaki fragments, following replacement of the RNA primers with DNA nucleotides by DNA polymerase δ. Okazaki fragments that are not ligated could cause double-strand-breaks, which cleaves the DNA. Since only a small number of double-strand breaks are tolerated, and only a small number can be repaired, enough ligation failures could be lethal to the cell. Further research implicates the supplementary role of Proliferating cell nuclear antigen (PCNA) to DNA Ligase I’s function of joining Okazaki fragments. When the PCNA binding site on DNA Ligase I is inactive, DNA Ligase I’s ability to connect Okazaki fragments was severely impaired. Thus, a proposed mechanism follows: after a PCNA-DNA polymerase δ complex synthesizes Okazaki fragments, the DNA polymerase δ is released. Then, DNA Ligase I binds to the PCNA, which is clamped to the nicks of the lagging strand, and catalyzes the formation of phosphodiester bonds.[12][13][14]
Flap endonuclease 1
Main article: Flap structure-specific endonuclease 1Flap endonuclease 1 (FEN1) is responsible for processing Okazaki fragments. It works with DNA polymerase to remove the RNA primer of an Okazaki fragment and can remove the 5’ ribonucleotide and 5’ flaps when DNA polymerase displaces the strands during lagging strand synthesis. The removal of these flaps involves a process called nick translation and creates a nick for ligation. Thus, FEN1’s function is necessary to Okazaki fragment maturation in forming a long continuous DNA strand. Likewise, during DNA base repair, the damaged nucleotide is displaced into a flap and subsequently removed by FEN1.[11][15]
Dna2 endonuclease
In the presence of a single stranded DNA-binding protein RPA, the DNA 5’ flaps become too long, and the nicks no longer fit as substrate for FEN1. This prevents the FEN1 from removing the 5′-flaps. Thus, Dna2’s role is to reduce the 3′ end of these fragments, making it possible for FEN1 to cut the flaps, and the Okazaki fragment maturation more efficient.[16]
Biological function
Although synthesis of the lagging strand involves only half the DNA in the nucleus, the complexity associated with processing Okazaki fragments is about twice that required to synthesize the leading strand. Even in small species such as yeast, Okazaki fragment maturation happens approximately a million times during a single round of DNA replication. Processing of Okazaki fragments is therefore very common and crucial for DNA replication and cell proliferation.
During this process, RNA and DNA primers are removed, allowing the Okazaki fragments to attach to the lagging DNA strand. While this process seems quite simple and repetitive, defects in Okazaki fragment maturation can cause DNA strand breakage which can cause varying forms of “chromosome aberrations”.[17] Severe defects of Okazaki fragment maturation may halt DNA replication and induce cell death. However, while subtle defects do not affect growth, they do result in future varying forms of genome instabilities. Based on the dangers associated with a failure in the DNA process, Okazaki fragments maintain our evolutionary development.
Okazaki fragments in prokaryotes and eukaryotes
DNA molecules in eukaryotes differ from the circular molecules of prokaryotes in that they are larger and usually have multiple origins of replication. This means that each eukaryotic chromosome is composed of many replicating units of DNA with multiple origins of replication. In comparison, the prokaryotic E. coli chromosome has only a single origin of replication. In eukaryotes, these replicating forks, which are numerous all along the DNA, form "bubbles" in the DNA during replication. The replication fork forms at a specific point called autonomously replicating sequences (ARS). Eukaryotes have a clamp loader complex and a six-unit clamp called the proliferating cell nuclear antigen.[18] The efficient movement of the replication fork also relies critically on the rapid placement of sliding clamps at newly primed sites on the lagging DNA strand by ATP-dependent clamp loader complexes. This means that the piecewise generation of Okazaki fragments can keep up with the continuous synthesis of DNA on the leading strand. These clamp loader complexes are characteristic of all eukaryotes and separate some of the minor differences in the synthesis of Okazaki fragments in prokaryotes and eukaryotes.[19] The lengths of Okazaki fragments in prokaryotes and eukaryotes are different as well. Prokaryotes have Okazaki fragments that are quite longer than those of eukaryotes. Eukaryotes typically have Okazaki fragments that are 100 to 200 nucleotides long, whereas prokaryotic E. Coli can be 2,000 nucleotides long. Okazaki fragments are also produced much quicker in prokaryotes since prokaryotic cells contain less genetic material. The complexity of the DNA in eukaryotes calls for a longer production time of the Okazaki fragments.
Uses in technology
Medical concepts associated with Okazaki fragments
 A slight mutation in the matched nucleotides can lead to chromosomal aberrations and unintentional genetic rearrangement.
A slight mutation in the matched nucleotides can lead to chromosomal aberrations and unintentional genetic rearrangement.
Although cells undergo multiple steps in order to ensure there are no mutations in the genetic sequence, sometimes specific deletions and other genetic changes during Okazaki fragment maturation go unnoticed. Because Okazaki fragments are the set of nucleotides for the lagging strand, any alteration including deletions, insertions, or duplications from the original strand can cause a mutation if it is not detected and fixed. Other causes of mutations include problems with the proteins that aid in DNA replication. For example, a mutation related to primase affects RNA primer removal and can make the DNA strand more fragile and susceptible to breaks. Another mutation concerns polymerase α, which impairs the editing of the Okazaki fragment sequence and incorporation of the protein into the genetic material. Both alterations can lead to chromosomal aberrations, unintentional genetic rearrangement, and a variety of cancers later in life.[17]
To test the effects of the protein mutations on living organisms, researchers genetically altered lab mice to be homozygous for another mutation in protein related to DNA replication, flap endonuclease 1, or FEN1. The results varied based on the specific gene alterations. Homozygous knockout mutant mice experienced a “failure of cell proliferation” and “early embryonic lethality” (27). Mice with mutation F343A and F344A (also known as FFAA) died directly after birth due to complications including pancytopenia and pulmonary hypoplasia. This is because the FFAA mutation keeps FEN1 from interacting with PCNA (proliferating cell nuclear antigen), consequently not allowing it to complete its purpose during Okazaki fragment maturation. Under careful observation, cells homozygous for FFAA FEN1 mutations seem to display only partial defects in maturation, meaning mice heterozygous for the mutation would be able to survive into adulthood, despite sustaining multiple small nicks in their genomes. Inevitably however, these nicks prevent future DNA replication because the break causes the replication fork to collapse and causes double strand breaks in the actual DNA sequence. In time, these nicks also cause full chromosome breaks, which could lead to severe mutations and cancers. Other mutations have been implemented with altered versions of Polymerase α, leading to similar results.[17]
Engineering concepts associated with Okazaki fragments
Due Okazaki fragment’s importance and purpose in DNA replication, bioengineers are using these pieces of DNA in their research. One study, published in Science on March 17, 2000, stated that the specific start and stop locations of bidirectional DNA synthesis at a human gene called Lamin B2 were discovered using DNA base exploration and information about Okazaki fragment synthesis. Based on the relative position of Okazaki fragments to the replication fork during lagging stand synthesis as well as the average length of Okazaki fragments during replication, the scientists were able to pinpoint nucleotide 3933 as the start of bidirectional synthesis.[20] The information was, “deriving from the linking to the initiated ori of four subsequent Okazaki fragments having lengths of 143, 144, 65, and 80 nucleotides from the first to the fourth, respectively”.
Another study investigated the formation of Okazaki fragments in wild-type bacteria cells. In strains of Escherichia coli, a mutation in the sof locus, a specific location of a gene on a chromosome, resulted in hyper recombination and short DNA strands. Scientists say that the mutated gene is defective, affecting an enzyme called deoxyuridinetriphosphate diphosphohydrolase (or dUTPase), which can no longer catalyze the hydrolysis of dUTP. This mutation also decreases the amount of dUTPase in the organism, somehow increasing the amount of uracil that is being incorporated into the DNA. Because uracil is not one of the four nucleotides associated with DNA (replaced by Thymine), it must be removed as an error by the excision-repair process and replaced. Rapid removal of uracil because of increased incorporation into DNA can lead to the accumulation of short DNA fragments, which may also lead to the creation of Okazaki fragments.[21] Learning how Okazaki fragments originate in bacteria such as E. coli allows scientists and engineers to better understand the process of DNA replication and the effects of pinpoint mutations.
Although many mutations associated with Okazaki fragments can result in cancer, studies have shown that the alteration of Okazaki fragments in nucleoside analogues such as cytarabine can lead to anti-cancer activities. For instance, cytarabine is an anti-leukemic agent that inhibits the replication of lagging stand DNA. When this agent was substituted for deoxycytidine during the replication of the SV-40 viral genome, the bending of the DNA duplex region (DDR), and the RNA-DNA hybrid duplex region (HDR) increased from 22 degrees to 41 degrees. This increased helical bending of Okazaki fragments may contribute to cytarabine's ability to inhibit the replication of lagging strand DNA and have anti-cancer properties.[22]
References
- ^ Goldstein, Elliott; Lewin, Benjamin; Jocelyn E. Krebs; Stephen T. Kilpatrick (2011). Lewin's genes X. Boston: Jones and Bartlett. p. 329. ISBN 0-7637-7992-X.
- ^ Sakabe K, Okazaki R (December 1966). "A unique property of the replicating region of chromosomal DNA". Biochimica et Biophysica Acta 129 (3): 651–54. PMID 5337977.
- ^ Okazaki R, Okazaki T, Sakabe K, Sugimoto K (June 1967). "Mechanism of DNA replication possible discontinuity of DNA chain growth". Japanese Journal of Medical Science & Biology 20 (3): 255–60. PMID 4861623.
- ^ An American scientist, by the last name Shandel, discovered this mechanism prior to Okazaki, however he was never accredited with the discovery since the head of his research team decided the discovery was an erroneous interpretation of test results.
- ^ a b Ogawa T, Okazaki T (1980). "Discontinuous DNA replication". Annual Review of Biochemistry 49: 421–57. doi:10.1146/annurev.bi.49.070180.002225. PMID 6250445. http://arjournals.annualreviews.org/doi/full/10.1146/annurev.bi.49.070180.002225?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed.
- ^ Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A (February 1968). "Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains". Proceedings of the National Academy of Sciences of the United States of America 59 (2): 598–605. doi:10.1073/pnas.59.2.598. PMC 224714. PMID 4967086. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=224714.
- ^ Sugimoto K, Okazaki T, Okazaki R (August 1968). "Mechanism of DNA chain growth, II. Accumulation of newly synthesized short chains in E. coli infected with ligase-defective T4 phages". Proceedings of the National Academy of Sciences of the United States of America 60 (4): 1356–62. doi:10.1073/pnas.60.4.1356. PMC 224926. PMID 4299945. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=224926.
- ^ Henry RA, Balakrishnan L, Ying-Lin ST, Campbell JL, Bambara RA (September 2010). "Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing". The Journal of Biological Chemistry 285 (37): 28496–505. doi:10.1074/jbc.M110.131870. PMID 20628185. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=20628185.
- ^ a b Pike JE, Henry RA, Burgers PM, Campbell JL, Bambara RA (December 2010). "An alternative pathway for Okazaki fragment processing: resolution of fold-back flaps by Pif1 helicase". The Journal of Biological Chemistry 285 (53): 41712–23. doi:10.1074/jbc.M110.146894. PMID 20959454. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=20959454.
- ^ Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM (February 2006). "DNA primase acts as a molecular brake in DNA replication". Nature 439 (7076): 621–4. doi:10.1038/nature04317. PMID 16452983.
- ^ a b Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA (April 2001). "The 3'-->5' exonuclease of DNA polymerase delta can substitute for the 5' flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability". Proceedings of the National Academy of Sciences of the United States of America 98 (9): 5122–7. doi:10.1073/pnas.091095198. PMC 33174. PMID 11309502. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=11309502.
- ^ a b Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM (January 2003). "Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3'-5'-exonuclease activities of Pol delta in the creation of a ligatable nick". The Journal of Biological Chemistry 278 (3): 1626–33. doi:10.1074/jbc.M209803200. PMID 12424237. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=12424237.
- ^ Levin DS, Bai W, Yao N, O'Donnell M, Tomkinson AE (November 1997). "An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining". Proceedings of the National Academy of Sciences of the United States of America 94 (24): 12863–8. doi:10.1073/pnas.94.24.12863. PMC 24229. PMID 9371766. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=9371766.
- ^ Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE (2000). "Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair". Current Biology : CB 10 (15): 919–22. doi:10.1016/S0960-9822(00)00619-9. PMID 10959839. http://linkinghub.elsevier.com/retrieve/pii/S0960-9822(00)00619-9.
- ^ Liu Y, Kao HI, Bambara RA (2004). "Flap endonuclease 1: a central component of DNA metabolism". Annual Review of Biochemistry 73: 589–615. doi:10.1146/annurev.biochem.73.012803.092453. PMID 15189154. http://arjournals.annualreviews.org/doi/full/10.1146/annurev.biochem.73.012803.092453?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed.
- ^ Ayyagari R, Gomes XV, Gordenin DA, Burgers PM (January 2003). "Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2". The Journal of Biological Chemistry 278 (3): 1618–25. doi:10.1074/jbc.M209801200. PMID 12424238. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=12424238.
- ^ a b c Zheng L, Shen B (February 2011). "Okazaki fragment maturation: nucleases take centre stage". Journal of Molecular Cell Biology 3 (1): 23–30. doi:10.1093/jmcb/mjq048. PMC 3030970. PMID 21278448. http://jmcb.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=21278448.
- ^ "Eukaryotic DNA Replication." Molecular-Plant-Biotechnology. multilab.biz, n.d.Web. 29 Mar. 2011.
- ^ Matsunaga F, Norais C, Forterre P, Myllykallio H (February 2003). "Identification of short 'eukaryotic' Okazaki fragments synthesized from a prokaryotic replication origin". EMBO Reports 4 (2): 154–8. doi:10.1038/sj.embor.embor732. PMC 1315830. PMID 12612604. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1315830.
- ^ Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A (March 2000). "Start sites of bidirectional DNA synthesis at the human lamin B2 origin". Science 287 (5460): 2023–6. doi:10.1126/science.287.5460.2023. PMID 10720330. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=10720330.
- ^ Tye BK, Nyman PO, Lehman IR, Hochhauser S, Weiss B (January 1977). "Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA". Proceedings of the National Academy of Sciences of the United States of America 74 (1): 154–7. doi:10.1073/pnas.74.1.154. PMC 393216. PMID 319455. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=393216.
- ^ Gmeiner WH, Konerding D, James TL (January 1999). "Effect of cytarabine on the NMR structure of a model Okazaki fragment from the SV40 genome". Biochemistry 38 (4): 1166–75. doi:10.1021/bi981702s. PMID 9930976.
- Notes
- Inman RB, Schnos M, Structure of branch points in replicating DNA: Presence of single-stranded connections in lambda DNA branch points. J. Mol Biol. 56:319-625, 1971.
- Thommes P, Hubscher U. Eukaryotic DNA replication. Enzymes and proteins acting at the fork. Eur. J. Biochem. 194(3):699-712, 1990.
External links
Categories:
Wikimedia Foundation. 2010.