- Double-stranded RNA viruses
-
Double-stranded RNA viruses 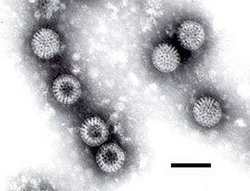
Electron micrograph of rotaviruses. The bar = 100 nm Virus classification Group: Group III (dsRNA) Families Birnaviridae
Chrysoviridae
Cystoviridae
Endornaviridae
Hypoviridae
Partitiviridae
Picobirnaviridae
Reoviridae
TotiviridaeDouble-stranded (ds) RNA viruses are a diverse group of viruses that vary widely in host range (humans, animals, plants, fungi, and bacteria), genome segment number (one to twelve), and virion organization (T-number, capsid layers, or turrets). Members of this group include the rotaviruses, known globally as a common cause of gastroenteritis in young children, and bluetongue virus, an economically important pathogen of cattle and sheep.
Of these families, the Reoviridae is the largest and most diverse in terms of host range.
In recent years virus particle assembly, virus-cell interactions, and viral pathogenesis, approaches for the development of novel antiviral strategies or agents can be designed.[1]
Contents
Taxa
Viruses with dsRNA genomes are currently grouped into a number of families, unassigned genera and species:
Families
- Birnaviridae
- Chrysoviridae
- Cystoviridae
- Endornaviridae
- Hypoviridae
- Partitiviridae
- Picobirnaviridae
- Reoviridae
- Totiviridae
Unassigned genera
Unassigned species
- Sclerotinia sclerotiorum debilitation associated virus
Notes on selected species
Reoviridae
Reoviridae are currently classified into nine genera. The genomes of these viruses consist of 10 to 12 segments of dsRNA, each generally encoding one protein. The mature virions are non-enveloped. Their capsids, formed by multiple proteins, have icosahedral symmetry and are arranged generally in concentric layers. A distinguishing feature of the dsRNA viruses, irrespective of their family association, is their ability to carry out transcription of the dsRNA segments, under appropriate conditions, within the capsid. In all these viruses, the enzymes required for endogenous transcription are thus part of the virion structure.[1]
Orthoreoviruses
The orthoreoviruses (reoviruses) are the prototypic members of the virus Reoviridae family and representative of the turreted members, which comprise about half the genera. Like other members of the family, the reoviruses are non-enveloped and characterized by concentric capsid shells that encapsidate a segmented dsRNA genome. In particular, reovirus has eight structural proteins and ten segments of dsRNA. A series of uncoating steps and conformational changes accompany cell entry and replication. High-resolution structures are known for almost all of the proteins of mammalian reovirus (MRV), which is the best-studied genotype. Electron cryo-microscopy (cryoEM) and X-ray crystallography have provided a wealth of structural information about two specific MRV strains, type 1 Lang (T1L) and type 3 Dearing (T3D).[2]
Cypovirus
The cytoplasmic polyhedrosis viruses (CPVs) form the genus Cypovirus of the family Reoviridae. CPVs are classified into 14 species based on the electrophoretic migration profiles of their genome segments. Cypovirus has only a single capsid shell, which is similar to the orthoreovirus inner core. CPV exhibits striking capsid stability and is fully capable of endogenous RNA transcription and processing. The overall folds of CPV proteins are similar to those of other reoviruses. However, CPV proteins have insertional domains and unique structures that contribute to their extensive intermolecular interactions. The CPV turret protein contains two methylase domains with a highly conserved helix-pair/β-sheet/helix-pair sandwich fold but lacks the β-barrel flap present in orthoreovirus λ2. The stacking of turret protein functional domains and the presence of constrictions and A spikes along the mRNA release pathway indicate a mechanism that uses pores and channels to regulate the highly coordinated steps of RNA transcription, processing, and release.[3]
Rotavirus
Rotavirus is the most common cause of acute gastroenteritis in infants and young children worldwide. This virus contains a dsRNA genome and is a member of the Reoviridae family. The genome of rotavirus consists of eleven segments of dsRNA. Each genome segment codes for one protein with the exception of segment 11, which codes for two proteins. Among the twelve proteins, six are structural and six are non-structural proteins.[4] It is a double-stranded RNA non-enveloped virus
Bluetongue virus
The members of Orbivirus genus within the Reoviridae family are arthropod-borne viruses and are responsible for high morbidity and mortality in ruminants. Bluetongue virus (BTV) which causes disease in livestock (sheep, goat, cattle) has been in the forefront of molecular studies for the last three decades and now represents the best understood orbivirus at the molecular and structural levels. BTV, like other members of the family, is a complex non-enveloped virus with seven structural proteins and a RNA genome consisting of 10 variously sized dsRNA segments.[5][6]
Phytoreoviruses
Phytoreoviruses are non-turreted reoviruses that are major agricultural pathogens, particularly in Asia. One member of this family, Rice Dwarf Virus (RDV), has been extensively studied by electron cryomicroscopy and x-ray crystallography. From these analyses, atomic models of the capsid proteins and a plausible model for capsid assembly have been derived. While the structural proteins of RDV share no sequence similarity to other proteins, their folds and the overall capsid structure are similar to those of other Reoviridae.[7]
The Yeast dsRNA Virus L-A
The L-A dsRNA virus of the yeast Saccharomyces cerevisiae has a single 4.6 kb genomic segment that encodes its major coat protein, Gag (76 kDa) and a Gag-Pol fusion protein (180 kDa) formed by a -1 ribosomal frameshift. L-A can support the replication and encapsidation in separate viral particles of any of several satellite dsRNAs, called M dsRNAs, each of which encodes a secreted protein toxin (the killer toxin) and immunity to that toxin. L-A and M are transmitted from cell to cell by the cytoplasmic mixing that occurs in the process of mating. Neither is naturally released from the cell or enters cells by other mechanisms, but the high frequency of yeast mating in nature results in the wide distribution of these viruses in natural isolates. Moreover, the structural and functional similarities with dsRNA viruses of mammals has made it useful to consider these entities as viruses.[8]
Infectious bursal disease virus
Infectious bursal disease virus (IBDV) is the best-characterized member of the family Birnaviridae. These viruses have bipartite dsRNA genomes enclosed in single-layered icosahedral capsids with T = 13l geometry. IBDV shares functional strategies and structural features with many other icosahedral dsRNA viruses, except that it lacks the T = 1 (or pseudo T = 2) core common to the Reoviridae, Cystoviridae, and Totiviridae. The IBDV capsid protein exhibits structural domains that show homology to those of the capsid proteins of some positive-sense single-stranded RNA viruses, such as the nodaviruses and tetraviruses, as well as the T = 13 capsid shell protein of the Reoviridae. The T = 13 shell of the IBDV capsid is formed by trimers of VP2, a protein generated by removal of the C-terminal domain from its precursor, pVP2. The trimming of pVP2 is performed on immature particles as part of the maturation process. The other major structural protein, VP3, is a multifunctional component lying under the T = 13 shell that influences the inherent structural polymorphism of pVP2. The virus-encoded RNA-dependent RNA polymerase, VP1, is incorporated into the capsid through its association with VP3. VP3 also interacts extensively with the viral dsRNA genome.[9]
dsRNA bacteriophage Φ6
Bacteriophage Φ6, is a member of the Cystoviridae family. It infects Pseudomonas bacteria (typically plant-pathogenic P. syringae). It has a three-part, segmented, double-stranded RNA genome, totalling ~13.5 kb in length. Φ6 and its relatives have a lipid membrane around their nucleocapsid, a rare trait among bacteriophages. It is a lytic phage, though under certain circumstances has been observed to display a delay in lysis which may be described as a "carrier state".[10]
Anti-virals
Since cells do not produce double-stranded RNA during normal nucleic acid metabolism, natural selection has favored the evolution of enzymes that destroy dsRNA on contact. The best known class of this type of enzymes is Dicer. It is hoped that broad-spectrum anti-virals could be synthesized that take advantage of this vulnerability of double-stranded RNA viruses.[11]
See also
Microbiology: Virus Components Viral life cycle Genetics Other Viral disease · Laboratory diagnosis of viral infections · Viral load · Viral quantification · Antiviral drug · Bacteriophage · Neurotropic virus · Oncovirus · History of virusesReferences
- ^ Dryden et al. (2008). "The Structure of Orthoreoviruses". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Zhou ZH (2008). "Cypovirus". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Jiang et al. (2008). "Rotavirus Structure". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Roy P (2008). "Structure and Function of Bluetongue Virus and its Proteins". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Mettenleiter TC and Sobrino F (editors). (2008). Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6 . http://www.horizonpress.com/avir.
- ^ Baker et al. (2008). "Structures of Phytoreoviruses". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Wickner et al. (2008). "The Yeast dsRNA Virus L-A Resembles Mammalian dsRNA Virus Cores". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Caston et al. (2008). "Infectious Bursal Disease Virus (IBDV)". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Koivunen et al. (2008). "Structure-Function Insights Into the RNA-Dependent RNA Polymerase of the dsRNA Bacteriophage Φ6". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9. http://www.horizonpress.com/rnav.
- ^ Rider, Todd H.; Zook, Christina E.; Boettcher, Tara L.; Wick, Scott T.; Pancoast, Jennifer S.; Zusman, Benjamin D. (2011). "Broad-Spectrum Antiviral Therapeutics". PLoS ONE 6 (7). doi:e22572. doi:10.1371/journal.pone.0022572. "...a new broad-spectrum antiviral approach, dubbed Double-stranded RNA (dsRNA) Activated Caspase Oligomerizer (DRACO)..."
External links
Baltimore (virus classification) DNA I: dsDNA viruses HerpesviralesUnassignedNLCDV: Asfarviridae · Iridoviridae · Marseilleviridae · Megaviridae · Mimiviridae · Phycodnaviridae · Poxviridae
nonenveloped: Adenoviridae · Papillomaviridae · Papovaviridae (obsolete) · Polyomaviridae
Ascoviridae · Baculoviridae · Corticoviridae · Fuselloviridae · Guttaviridae · Lipothrixviridae · Nimaviridae · Plasmaviridae · Rudiviridae · TectiviridaeII: ssDNA viruses non-enveloped: Parvoviridae (Parvovirus B19)
ungrouped: Circoviridae · Geminiviridae · Inoviridae · Microviridae · NanoviridaeRNA III: dsRNA viruses Birnaviridae · Chrysoviridae · Cystoviridae · Hypoviridae · Partitiviridae · Reoviridae (Rotavirus) · TotiviridaeIV: (+)ssRNA viruses (primarily icosahedral) PicornaviralesDicistroviridae · Iflaviridae · Marnaviridae · Picornaviridae (Enterovirus, Rhinovirus) · SecoviridaeTymoviralesUnassignedAstroviridae · Barnaviridae · Bromoviridae · Caliciviridae · Closteroviridae · Comoviridae · Flaviviridae · Flexiviridae · Leviviridae · Luteoviridae · Narnaviridae · Nodaviridae · Potyviridae · Sequiviridae · Tetraviridae · Togaviridae (Rubella virus) · TombusviridaeV: (-)ssRNA viruses (primarily helical) RT VI: ssRNA-RT viruses VII: dsDNA-RT viruses Categories:- RNA
- Molecular biology
- Animal virology
- RNA viruses
Wikimedia Foundation. 2010.
