- Antigenic shift
-
Not to be confused with antigenic drift or genetic drift.
Antigenic shift is the process by which two or more different strains of a virus, or strains of two or more different viruses, combine to form a new subtype having a mixture of the surface antigens of the two or more original strains. The term is often applied specifically to influenza, as that is the best-known example, but the process is also known to occur with other viruses, such as visna virus in sheep.[1] Antigenic shift is a specific case of reassortment or viral shift that confers a phenotypic change.
In terms of virology, the marine ecosystem has been largely unstudied, but due to its extraordinary volume, high viral density (100 million viruses per mL in coastal waters, 3 million per mL in the deep sea)[2] and high cell lysing rate (as high as 20% on average), marine viruses' antigenic shift and genetic recombination rates must be quite high.[3] This is most striking when one considers that the coevolution of prokaryotes and viruses in the aquatic environment has been going on since before eukaryotes appeared on earth.
Antigenic shift is contrasted with antigenic drift, which is the natural mutation over time of known strains of influenza (or other things, in a more general sense) which may lead to a loss of immunity, or in vaccine mismatch. Antigenic drift occurs in all types of influenza including influenzavirus A, influenza B and influenza C. Antigenic shift, however, occurs only in influenzavirus A because it infects more than just humans.[4] Affected species include other mammals and birds, giving influenza A the opportunity for a major reorganization of surface antigens. Influenza B and C principally infect humans, minimizing the chance that a reassortment will change its phenotype drastically.[5]
Antigenic shift is important for the emergence of new viral pathogens as it is a pathway that viruses may follow to enter a new niche. It could occur with primate viruses and may be a factor for the appearance of new viruses in the human species such as HIV. Due to the structure of its genome, HIV does not undergo reassortment, but it does recombine freely and via superinfection HIV can produce recombinant HIV strains that differ significantly from their ancestors.
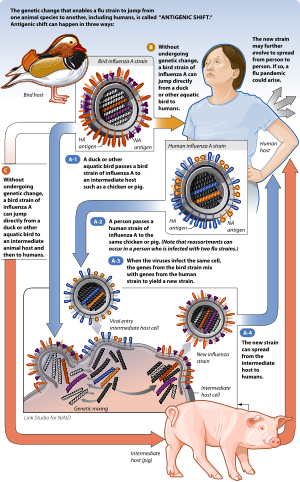
Flu strains are named after their types of hemagglutinin and neuraminidase surface proteins, so they will be called, for example, H3N2 for type-3 hemagglutinin and type-2 neuraminidase. When two [[different strains of influenza infect the same cell simultaneously, their protein capsids and lipid envelopes are removed, exposing their RNA, which is then transcribed to mRNA. The host cell then forms new viruses that combine their antigens; for example, H3N2 and H5N1 can form H5N2 this way. Because the human immune system has difficulty recognizing the new influenza strain, it may be highly dangerous. Influenza viruses which have undergone antigenic shift have caused the Asian Flu pandemic of 1957, the Hong Kong Flu pandemic of 1968, and the Swine Flu scare of 1976. Until recently, such combinations were believed to have caused the infamous Spanish Flu outbreak of 1918 which killed 40~100 million people worldwide; however more recent research suggests the 1918 pandemic was caused by the antigenic drift of a fully avian virus to a form that could infect humans efficiently.[6][7] One increasingly worrying situation is the possible antigenic shift between avian influenza and human influenza. This antigenic shift could cause the formation of a highly virulent virus.
Contents
Role in transmission of influenza viruses from animals to people
Influenza A viruses are found in many different animals, including ducks, chickens, pigs, whales, horses, and seals.[5] Influenza B viruses circulate widely principally among humans, though it has recently been found in seals.[8]
There are 16 different hemagglutinin subtypes and 9 different neuraminidase subtypes, all of which have been found among influenza A viruses in wild birds. Wild birds are the primary natural reservoir for all subtypes of influenza A viruses and are thought to be the source of influenza A viruses in all other animals.[4] Most influenza viruses cause asymptomatic or mild infection in birds; however, the range of symptoms in birds varies greatly depending on the strain of virus. Infection with certain avian influenza A viruses (for example, some strains of H5 and H7 viruses) can cause widespread disease and death among some species of wild and especially domestic birds such as chickens and turkeys.
Pigs can be infected with both human and avian influenza viruses in addition to swine influenza viruses. Infected pigs get symptoms similar to humans, such as cough, fever, and runny nose. Because pigs are susceptible to avian, human and swine influenza viruses, they potentially may be infected with influenza viruses from different species (e.g., ducks and humans) at the same time. If this happens, it is possible for the genes of these viruses to mix and create a new virus.
For example, if a pig was infected with a human influenza virus and an avian influenza virus at the same time, an antigenic shift could occur, producing a new virus that had most of the genes from the human virus, but a hemagglutinin or neuraminidase from the avian virus. The resulting new virus would likely be able to infect humans and spread from person to person, but it would have surface proteins (hemagglutinin and/or neuraminidase) not previously seen in influenza viruses that infect humans, and therefore to which most people have little or no immune protection. If this new virus causes illness in people and can be transmitted easily from person to person, an influenza pandemic can occur.[5] The most recent 2009 H1N1 outbreak was a result of antigenic shift and reassortment between human, avian, and swine viruses. [9]
See also
Notes
- ^ Narayan, O; Griffin, DE; Chase, J (1977). "Antigenic shift of visna virus in persistently infected sheep". Science.) 197 (4301): 376–378. doi:10.1126/science.195339. PMID 195339.)
- ^ Denny, How the Ocean Works: An Introduction to Oceanography (2008). How the Ocean Works (illustrated ed.). Princeton University Press. ISBN 069112647X, 9780691126470.)
- ^ Suttle, CA (2007). "Marine viruses — major players in the global ecosystem". Nature. 5 (10): 801–812. doi:10.1038/nrmicro1750. PMID 17853907.
- ^ a b Treanor, John (2004-01-15). "Influenza vaccine--outmaneuvering antigenic shift and drift". New England Journal of Medicine 350 (3): 218–220. doi:10.1056/NEJMp038238. PMID 14724300. http://content.nejm.org/cgi/content/full/350/3/218. Retrieved 2008-01-07.
- ^ a b c Zambon, Maria C. (November 1999). "Epidemiology and pathogenesis of influenza" (PDF). Journal of Antimicrobial Chemotherapy 44 (Supp B): 3–9. doi:10.1093/jac/44.suppl_2.3. PMID 10877456. http://jac.oxfordjournals.org/cgi/reprint/44/suppl_2/3.pdf. Retrieved 2008-01-09.
- ^ Aoki, FY; Sitar, DS (January 1988). "Clinical pharmacokinetics of amantadine hydrochloride". Clinical Pharmacokinetics 14 (1): 35–51. doi:10.2165/00003088-198814010-00003. PMID 3280212.
- ^ Johnson, NP; Mueller, J (2002 Spring). "Updating the accounts: global mortality of the 1918-1920 "Spanish" influenza pandemic". Bulletin of the History of Medicine. 76 (1): 105–115. doi:10.1353/bhm.2002.0022. PMID 11875246.
- ^ Carrington, Damian (2000-05-11). "Seals pose influenza threat". BBC. http://news.bbc.co.uk/1/hi/sci/tech/744945.stm.
- ^ Smith, G. J. D.; D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. M. Peiris, Y. Guan, A. Rambaut (2002). "Dating the emergence of pandemic influenza viruses". Proceedings of the National Academy of Sciences of the USA 106: 11709–11712.
Further reading
- "The biology of influenza viruses". Vaccine 26 Suppl 4: D49–53. September 2008. PMC 3074182. PMID 19230160. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=3074182.
External links
Microbiology: Virus Components Viral life cycle Genetics Other Viral disease · Laboratory diagnosis of viral infections · Viral load · Viral quantification · Antiviral drug · Bacteriophage · Neurotropic virus · Oncovirus · History of virusesCategories:
Wikimedia Foundation. 2010.