- DLC1
-
Deleted in Liver Cancer 1 also known as DLC1 and StAR-related lipid transfer protein 12 (STARD12) is a protein which in humans is encoded by the DLC1 gene.[1][2]
This gene is deleted in the primary tumor of hepatocellular carcinoma. It maps to 8p22-p21.3, a region frequently deleted in solid tumors. It is suggested that this gene is a candidate tumor suppressor gene for human liver cancer, as well as for prostate, lung, colorectal, and breast cancers.[3]
Contents
Gene
The human DLC1 gene is located on the short arm of chromosome 8 (8p21.3-22), within a region that frequently undergoes loss of heterozygosity by either genomic deletion or epigenetic silencing mechanisms in several types of solid cancers.[4] The gene contains 14 exons and produces an mRNA transcript that is 6.3 kb in length; the second AUG present in the open reading frame is the major translational start site, and produces a polypeptide which is 1091 amino acids long.[5]
The promoter region of the DLC1 gene contains a CpG island containing several CpG sites which can be methylated to promote gene silencing and prevent transcription.[6]
DLC1 is frequently inactivated in human hepatocellular carcinoma, as well as some nasopharyngeal, lung, breast, prostate, kidney, colon, uterine, ovarian, and gastric cancers.[7]
Protein structure and localization
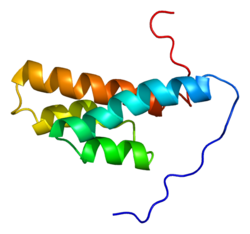
The DLC1 protein contains four major functional domains: an N-terminal sterile α motif (SAM), a serine-rich (SR) region, a Rho-GAP domain, and a C-terminal steroidogenic acute regulatory protein related lipid-transfer (START) domain.[5] DLC1 is localized to focal adhesions located at the periphery of cells.
SAM domain
The SAM domain (stretching from amino acids 11-78) is believed to be involved in protein-protein interactions. The exact function of the DLC1 SAM domain has not yet been determined.[5]
SR region
The relatively unstructured and unconserved SR region (amino acids 86-638) contains a focal adhesion targeting (FAT) domain,[7] including a tyrosine residue at position 442, which interacts with SH2 domains of tensin1[8] and cten.[9] These interactions allow DLC1 to co-localize along with these proteins to focal adhesions at the periphery of the cell, where it is able to carry out its function as a Rho-GAP protein.
Rho-GAP domain
The highly conserved Rho-GAP domain (amino acids 639-847) functions to enhance the GTPase activity of the Rho-GTPase proteins RhoA and Cdc42, promoting the hydrolysis of their bound GTP to GDP and thus “shutting off” these proteins. DLC1 contains a conserved “arginine finger” arginine residue at position 677, which is located within the active site of the protein and is essential for catalyzation of the GTP hydrolysis[5]. Rho-GTPases are involved in regulating cell morphology (through cytoskeletal organization) and migration (through focal adhesion formation).[10]
START domain
The START domain (amino acids 878-1081) contains a β-sheet which forms a hydrophobic tunnel held in place by α-helices.[5] This region interacts with phospholipase C-δ1 (PLCδ1) and activates its ability to hydrolyze the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) into diacylglycerol (DAG) and inositol 1,4,5-triphosphate (IP3), which in turn activates protein kinase C (PKC) and increases intracellular calcium ion concentration, which regulates the actin cytoskeleton.[7] In addition, hydrolysis of PIP2 releases actin regulatory proteins assembled at PIP2 molecules on the membrane and allows them to promote the disassembly of actin filaments.[5] The C-terminus of DLC1 is also known to interact with caveolin-1, although the biological significance of this interaction has not yet been discovered.[5]
Role in embryogenesis
The mouse homologue of DLC1 was required during embryogenesis. While mice heterozygous for the dlc1 gene showed no physical abnormalities, mouse embryos which are homozygous negative for dlc-1 were not able to progress past ten and a half days gestation.[11] Further analysis of the embryos revealed that they had defects in several organs, including the brain, heart, and placenta. In addition, cells of the DLC1-/- embryos had few long actin fibers (indicating that their cytoskeletal organization was impaired) and fewer focal adhesions than those of normal DLC1 expressing cells.[11]
Significance in cancer
As previously mentioned, the dlc1 gene is found to be deleted or down-regulated in several solid cancers, including human liver, non-small cell lung, nasopharyngeal, breast, prostate, kidney, colon, uterine, ovarian, and stomach cancers.[7] It acts as a tumor suppressor gene to inhibit cell growth and proliferation as well as induce apoptosis when a cell is under stress. DLC1 is also involved in the formation of focal adhesions, so loss of DLC1 leads to reduced cell adhesion and increased metastatic potential of cells.
Tumor suppressor gene activity
DLC1 expression is frequently lost in tumor cells, resulting in constitutive activation of the RhoGTPases RhoA and Cdc42. This results in increased cell growth and proliferation, changes in cell morphology, and inhibition of apoptosis.
A tumor suppressor gene is a gene whose protein product acts to prevent cells from proliferating at inappropriate times, or to induce apoptosis of cells which are damaged beyond repair.
The loss of heterozygosity of DLC1 results when one copy of the gene is deleted or inactivated, but because of the presence of a second functional copy of the gene, no phenotypic changes are observed. However, if this second copy is then deleted or inactivated, the protein is no longer able to be expressed, and changes in cellular phenotype and tumorigenesis may result. These observations are consistent with the tumor suppression properties of DLC1.
The main function of DLC1 is its Rho-GAP activity: its ability to enhance activated GTP-bound Rho-GTPases' (specifically, RhoA and Cdc42) intrinsic ability to convert their GTP into GDP, thus rendering them inactive. RhoGTPases are members of the Ras superfamily, and are involved in actin cytoskeleton organization and cell adhesion.[12] The activity of RhoA regulates the formation of actin stress fibers and focal adhesions - complexes of many proteins located at the termini of actin stress fibers which link the actin cytoskeleton with integrin extracellular matrix receptors. Therefore, when RhoA is inactive, the actin cytoskeletal filaments are unable to form and cell morphology changes, resulting in a default round shape.[10] In addition, focal adhesion formation is inhibited and cells are not well attached to the extracellular matrix and neighbouring cells[5], thus allowing them to detach and metastasize more readily.
The Rho-GTPase Cdc42 is involved in regulation of the cell cycle and preventing inappropriate cell division.[13] Constitutive activation of Cdc42 due to the absence of RhoGAP proteins such as DLC1 will contribute to the continual repetition of the cell cycle, resulting in uncontrolled cell growth and proliferation.
The addition of DLC1 to tumor cells lines which are deficient in DLC1 expression reduces the RhoA-GTP levels in the cells, which in turn promotes the disassembly of actin stress fibers and cause cells to adopt a rounded morphology.[10] Overexpression of DLC1 also results in inhibited cell growth, proliferation, tumor formation, migration, and increased apoptosis.[12]
Involvement in signalling pathways
DLC1 is involved in the phophoinositide and insulin signaling cascades.
As mentioned, the C-terminal START domain of DLC1 is involved in phosphoinositide signaling[5]: it is able to interact with phospholipase C-δ1 (PLC- δ1), thereby stimulating it to hydrolyze phosphatidylinositol 4,5-bisphosphate (PIP2) into the second messengers inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 causes calcium to be released from vesicles into the cytoplasm, which in turn regulates proteins which are sensitive to high calcium concentrations. DAG activates protein kinase C (PKC) and triggers a cascade of intracellular signals.
DLC1 may have additional role in insulin signaling, as the presence of insulin results in the phosphorylation of the serine residue at position 329 (within the SR region) on DLC1 by protein kinase B (PKB) aka AKT,[14] although the significance and function of this phosphorylation is as yet unknown.
Apoptosis
DLC1 is responsible for inducing programmed cell death by at least two mechanisms: caspase-3-mediated apoptosis and Bcl-2 activated mitochondrial-mediated apoptosis.
The process of apoptosis, or programmed cell death, allows cells which are stressed or damaged to die in a controlled and contained manner. Experiments have shown that DLC1 expression initiates a signaling cascade which cleaves the precursor protein procaspase-3 into caspase-3, thereby allowing it to induce caspase-3-mediated apoptosis.[12][15] Therefore, in the absence of DLC1, apoptosis of cells which are proliferating and passing through the cell cycle uncontrollably is significantly reduced.[12] These cells are unable to destroy themselves, and therefore continue to proliferate and form tumors.
DLC1 also performs a second pro-apoptotic function: it reduces cellular levels of the anti-apoptotic protein Bcl-2.[12] Mitochondrial-mediated apoptosis occurs when the ratio of the pro-apoptotic protein Bax and Bcl-2 is high; therefore, a reduction in Bcl-2 level will lead to an increase in the Bax/Bcl-2 ratio and induce mitochondrial-mediated apoptosis. In tumor cells which are not expressing DLC1, Bcl-2 levels remain high and the ration of Bax/Bcl-2 low, so apoptosis is inhibited.
The detailed pathways by which DLC1 results in the cleavage of procaspase-3 and decrease in Bcl-2 levels require further investigation.
Genomic instability
Current research does not suggest that DLC1 plays a role in destabilizing the genome and making it more susceptible to chromosomal rearrangements or gene mutations.
Hormonal regulation
DLC1 is known to be upregulated by at least two hormones: progesterone and peroxisome proliferators.
In ovarian cancers, DLC-1 expression is upregulated by the steroid hormone progesterone.[15] Gene profiling studies have shown that the addition of progesterone to ovarian cancer cell lines results in an increase in the expression of DLC1, which in turn results in growth inhibition, decreased cell motility, and increased caspase-3-mediated apoptosis.[15]
Lung cancer cells also increase DLC1 expression in response to peroxisome proliferator-activated receptor γ (PPARγ) activators.[16] PPARγ is a steroid hormone receptor which inhibits cellular growth of several epithelial cancers.
Role in migration and metastasis
In HCC, loss of DLC1 decreases focal adhesion turnover and allows cells to detach from primary tumors. In breast cancers, loss of DLC1 prevents cells from dividing and colonizing a new secondary tumor site.
DLC1 is downregulated in hepatocellular carcinoma cell lines, which, through the inactivation of Rho-GTPases, results in anchorage-independent growth in a semi-solid medium (soft agar), indicating that these cells are not held fast to their neighbors and can detach and are able to metastasize relatively easily.[10] Expression of DLC1 in hepatocellular carcinoma cells resulted in dephosphorylation of tyrosine residues on the molecule focal adhesion kinase (FAK), which results in the disassociation of focal adhesion complexes which are required for cell adhesion; therefore, dephosphorylation of FAK ultimately leads to an increase in focal adhesion turnover and cellular adhesion, and inhibition of cell migration.[10]
Furthermore in breast cancer cells, DLC1 functions as a metastasis-suppressor gene by inhibiting colonization of a secondary tumor site. Expression of DLC1 inhibited colonization ability by preventing any cells which were able to detach from the primary breast tumor and migrate to a secondary site from initiating division in the microenvironment of a new organ.[17]
Angiogenesis
As of 2010, current research indicates DLC1 negatively regulates angiogenesis in a paracrine fashion. This is by upregulation of VEGF mediated through the epidermal growth factor receptor (EGFR)-MAP/ERK Kinase (MEK)- hypoxia inducible factor 1 (HIF1) pathway.[18]
Epigenetic silencing
DLC1 expression is downregulated by both promoter hypermethylation and histone acetylation.
In hepatocellular carcinomas, the dlc1 gene is not always deleted, and can be detected in the tumor cells using PCR,[19] indicating that gene silencing through epigenetic mechanisms must also play an important role in downregulating DLC1 expression. They also demonstrated that the CpG island in the promoter region of the dlc1 gene is hypermethylated due to the action of DNA methyltransferase enzymes in hepatocellular carcinoma tumors[19], thus preventing the cells’ RNA polymerase and other transcriptional machinery from binding to the promoter an initiating transcription. This result was also verified in gastric cancer cells,[6] prostate cancer cells,[4] and other cancer cell lines with reduced DLC1 expression.
In addition, treatment of DLC1 downregulated tumor cell lines with a histone deacetylase inhibitor prevents histone deacetylase (HDAC) enzymes from removing acetyl groups from specific histones.[4] DNA is wrapped tightly around acetylated histones, thus preventing the transcriptional machinery from accessing the dlc1 gene, which is hidden within tightly packaged chromatin, and transcribing it into mRNA.
One hypothesis states that the activity of HDAC in the CpG region of the dlc1 gene promotes its silencing through interaction between the DNA and acetylated histone proteins. Following this, histone methyltransferases add methyl groups to the tail of histones (specifically, histone H3), which allows DNA methyltransferases to methylate the CpG’s of the dlc1 promoter itself, promoting the tight chromatin packaging which prevents transcription.[20]
Drug discovery and future therapies
The genomic deletion or downregulation of DLC1 expression in early tumors could serve as an indicator for future cancer progression and spread.[5]
Research into therapies for cancers with reduced levels of DLC1 expression due to epigenetic silencing could provide insight into the efficiency of epigenetic regulating molecules. For example, Zebularine, a demethylating agent, could be used to remove the methyl groups from the CpGs of the dlc1 promoter, thus increasing expression of DLC1 and helping to block tumor cell proliferation and metastasis. In addition, histone deacetylase inhibitors could potentially be used to prevent deacetylation of histones and loosen up the chromatin structure, thereby allowing RNA polymerase and other transcriptional proteins to reach the DNA and allow transcription to occur.[6]
Natural dietary flavones, found in parsley, celery, and citrus peels, reactivate DLC1 expression in breast cancer cell lines which have decreased DLC1 expression due to promoter hypermethylation, and may potentially be used as an anti-cancer agent for prevention and therapy of breast and other DLC1 downregulated cancers.[21]
References
- ^ Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC (May 1998). "Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP". Cancer Res. 58 (10): 2196–9. PMID 9605766. http://cancerres.aacrjournals.org/cgi/content/abstract/58/10/2196.
- ^ Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O (December 2000). "Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Res. 7 (6): 347–55. doi:10.1093/dnares/7.6.347. PMID 11214970. http://dnaresearch.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=11214970.
- ^ "Entrez Gene: DLC1". http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=10395.
- ^ a b c Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC (Mar 2006). "Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications". Clin Cancer Res. 12 (5): 1412–9. doi:10.1158/1078-0432.CCR-05-1906. PMID 16533763.
- ^ a b c d e f g h i j Durkin ME, Yuan BZ, Zhou X, et al. (2007). "DLC-1:a Rho GTPase-activating protein and tumour suppressor". J Cell Mol Med. 11 (5): 1185–207. doi:10.1111/j.1582-4934.2007.00098.x. PMID 17979893.
- ^ a b c Kim TY, Jong HS, Song SH, et al. (Jun 2003). "Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells". Oncogene 22 (25): 3943–51. doi:10.1038/sj.onc.1206573. PMID 12813468.
- ^ a b c d Liao YC, Lo SH (2008). "Deleted in liver cancer-1 (DLC-1): a tumor suppressor not just for liver". Int J Biochem Cell Biol. 40 (5): 843–7. doi:10.1016/j.biocel.2007.04.008. PMC 2323245. PMID 17521951. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2323245.
- ^ Qian X, Li G, Asmussen HK, et al. (May 2007). "Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities". Proc Natl Acad Sci USA. 104 (21): 9012–7. doi:10.1073/pnas.0703033104. PMC 1868654. PMID 17517630. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1868654.
- ^ Liao YC, Si L, deVere White RW, Lo SH (Jan 2007). "The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1". J Cell Biol. 176 (1): 43–9. doi:10.1083/jcb.200608015. PMC 2063623. PMID 17190795. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2063623.
- ^ a b c d e Kim TY, Lee JW, Kim HP, et al. (Mar 2007). "DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma". Biochem Biophys Res Commun. 355 (1): 72–7. doi:10.1016/j.bbrc.2007.01.121. PMID 17292327.
- ^ a b Durkin ME, Avner MR, Huh CG, Yuan BZ, Thorgeirsson SS, Popescu NC (Feb 2005). "DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development". FEBS Lett. 579 (5): 1191–6. doi:10.1016/j.febslet.2004.12.090. PMID 15710412.
- ^ a b c d e Zhou X, Thorgeirsson SS, Popescu NC (Feb 2004). "Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells". Oncogene 23 (6): 1308–13. doi:10.1038/sj.onc.1207246. PMID 14647417.
- ^ Fidyk N, Wang JB, Cerione RA (Jun 2006). "Influencing cellular transformation by modulating the rates of GTP hydrolysis by Cdc42". Biochemistry 45 (25): 7750–62. doi:10.1021/bi060365h. PMID 16784226.
- ^ Hers I, Wherlock M, Homma Y, Yagisawa H, Tavaré JM (Feb 2006). "Identification of p122RhoGAP (deleted in liver cancer-1) Serine 322 as a substrate for protein kinase B and ribosomal S6 kinase in insulin-stimulated cells". J Biol Chem. 281 (8): 4762–70. doi:10.1074/jbc.M511008200. PMID 16338927.
- ^ a b c Syed V, Mukherjee K, Lyons-Weiler J, et al. (Mar 2005). "Identification of ATF-3, caveolin-1, DLC-1, and NM23-H2 as putative antitumorigenic, progesterone-regulated genes for ovarian cancer cells by gene profiling". Oncogene 24 (10): 1774–87. doi:10.1038/sj.onc.1207991. PMID 15674352.
- ^ Grommes C, Landreth GE, Heneka MT (Jul 2004). "Antineoplastic effects of peroxisome proliferator-activated receptor gamma agonists". Lancet Oncol. 5 (7): 419–29. doi:10.1016/S1470-2045(04)01509-8. PMID 15231248.
- ^ Goodison S, Yuan J, Sloan D, et al. (Jul 2005). "The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells". Cancer Res. 65 (14): 6042–53. doi:10.1158/0008-5472.CAN-04-3043. PMC 1360170. PMID 16024604. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1360170.
- ^ Shih YP, Liao YC, Lin Y, Lo SH (2010). "DLC1 negatively regulates angiogenesis in a paracrine fashion". Cancer research 70 (21): 8270–5. doi:10.1158/0008-5472.CAN-10-1174. PMC 2970702. PMID 20861185. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2970702.
- ^ a b Wong CM, Lee JM, Ching YP, Jin DY, Ng IO (Nov 2003). "Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma". Cancer Res. 63 (22): 7646–51. PMID 14633684. http://cancerres.aacrjournals.org/cgi/pmidlookup?view=long&pmid=14633684.
- ^ Geiman TM, Robertson KD (2002). "Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together?". J Cell Biochem. 87 (2): 117–25. doi:10.1002/jcb.10286. PMID 12244565.
- ^ Ullmannova V, Popescu NC (2007). "Inhibition of cell proliferation, induction of apoptosis, reactivation of DLC1, and modulation of other gene expression by dietary flavone in breast cancer cell lines". Cancer Detect Prev. 31 (2): 110–8. doi:10.1016/j.cdp.2007.02.005. PMC 1950447. PMID 17418982. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1950447.
External links
PDB gallery Categories:- Human proteins
Wikimedia Foundation. 2010.