- Molecular self-assembly
-
Molecular self-assembly is the process by which molecules adopt a defined arrangement without guidance or management from an outside source. There are two types of self-assembly, intramolecular self-assembly and intermolecular self-assembly. Most often the term molecular self-assembly refers to intermolecular self-assembly, while the intramolecular analog is more commonly called folding.
Contents
Supramolecular systems
Molecular self-assembly is a key concept in supramolecular chemistry[2][3][4] since assembly of the molecules is directed through noncovalent interactions (e.g., hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π-π interactions, and/or electrostatic) as well as electromagnetic interactions. Common examples include the formation of micelles, vesicles, liquid crystal phases, and Langmuir monolayers by surfactant molecules.[5] Further examples of supramolecular assemblies demonstrate that a variety of different shapes and sizes can be obtained using molecular self-assembly.[6]
Molecular self-assembly has allowed the construction of challenging molecular topologies. An example are Borromean rings, interlocking rings wherein removal of one ring unlocks each of the other rings. DNA has been used to prepare a molecular analog of Borromean rings.[7] More recently, a similar structure has been prepared using non-biological building blocks.[8]
Biological systems
Molecular self-assembly is crucial to the function of cells. It is exhibited in the self-assembly of lipids to form the membrane, the formation of double helical DNA through hydrogen bonding of the individual strands, and the assembly of proteins to form quaternary structures. Molecular self-assembly of incorrectly folded proteins into insoluble amyloid fibers is responsible for infectious prion-related neurodegenerative diseases.
Nanotechnology
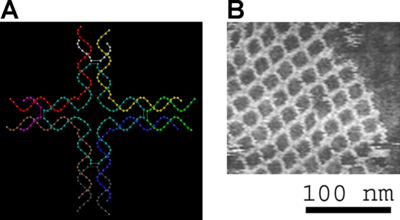 The DNA structure at left (schematic shown) will self-assemble into the structure visualized by atomic force microscopy at right. Image from Strong.[9]
The DNA structure at left (schematic shown) will self-assemble into the structure visualized by atomic force microscopy at right. Image from Strong.[9]
Molecular self-assembly is an important aspect of bottom-up approaches to nanotechnology. Using molecular self-assembly the final (desired) structure is programmed in the shape and functional groups of the molecules. Self-assembly is referred to as a 'bottom-up' manufacturing technique in contrast to a 'top-down' technique such as lithography where the desired final structure is carved from a larger block of matter. In the speculative vision of molecular nanotechnology, microchips of the future might be made by molecular self-assembly. An advantage to constructing nanostructure using molecular self-assembly for biological materials is that they will degrade back into individual molecules that can be broken down by the body.
DNA nanotechnology
Main article: DNA nanotechnologyDNA nanotechnology is an area of current research that uses the bottom-up, self-assembly approach for nanotechnological goals. DNA nanotechnology uses the unique molecular recognition properties of DNA and other nucleic acids to create self-assembling branched DNA complexes with useful properties.[10] DNA is thus used as a structural material rather than as a carrier of biological information, to make structures such as two-dimensional periodic lattices (both tile-based as well as using the "DNA origami" method) and three-dimensional structures in the shapes of polyhedra.[11] These DNA structures have also been used to template the assembly of other molecules such as gold nanoparticles[12] and streptavidin proteins.[13]
Two-dimensional monolayers
Main article: MonolayerThe spontaneous assembly of a single layer of molecules (i.e. monolayer thick) at interfaces is usually referred to as two-dimensional self-assembly. Early direct proofs showing that molecules can assembly into higher-order architectures at solid interfaces came with the development of scanning tunneling microscopy and shortly thereafter.[14] Eventually two strategies became popular for the self-assembly of 2D architectures, namely self-assembly following ultra-high-vacuum deposition and annealing and self-assembly at the solid-liquid interface.[15] The design of molecules and conditions leading to the formation of highly-crystalline architectures is considered today a form of 2D crystal engineering at the nanoscopic scale.
See also
References
- ^ F. H. Beijer, H. Kooijman, A. L. Spek, R. P. Sijbesma & E. W. Meijer (1998). "Self-Complementarity Achieved through Quadruple Hydrogen Bonding". Angew. Chem. Int. Ed. 37 (1-2): 75–78. doi:10.1002/(SICI)1521-3773(19980202)37:1/2<75::AID-ANIE75>3.0.CO;2-R.
- ^ J.-M. Lehn (1988). "Perspectives in Supramolecular Chemistry-From Molecular Recognition towards Molecular Information Processing and Self-Organization". Angew. Chem. Int. Ed. Engl. 27 (11): 89–121. doi:10.1002/anie.198800891.
- ^ J.-M. Lehn (1990). "Supramolecular Chemistry-Scope and Perspectives: Molecules, Supermolecules, and Molecular Devices (Nobel Lecture)". Angew. Chem. Int. Ed. Engl. 29 (11): 1304–1319. doi:10.1002/anie.199013041.
- ^ Lehn, J.-M.. Supramolecular Chemistry: Concepts and Perspectives. Wiley-VCH. ISBN 978-3-527-29311-7.
- ^ Rosen, Milton J. (2004). Surfactants and interfacial phenomena. Hoboken, NJ: Wiley-Interscience. ISBN 978-0-471-47818-8.
- ^ Ariga, Katsuhiko; Hill, Jonathan P; Lee, Michael V; Vinu, Ajayan; Charvet, Richard; Acharya, Somobrata (2008). "Challenges and breakthroughs in recent research on self-assembly" (free-download review). Science and Technology of Advanced Materials 9: 014109. doi:10.1088/1468-6996/9/1/014109.
- ^ C. Mao, W. Sun & N. C. Seeman (1997). "Assembly of Borromean rings from DNA". Nature 386 (6621): 137–138. doi:10.1038/386137b0. PMID 9062186
- ^ K. S. Chichak, S. J. Cantrill, A. R. Pease, S.-H. Chen, G. W. V. Cave, J. L. Atwood & J. F. Stoddart (2004). "Molecular Borromean Rings". Science 304 (5675): 1308–1312. doi:10.1126/science.1096914. PMID 15166376
- ^ M. Strong (2004). "Protein Nanomachines". PLoS Biol. 2 (3): e73-e74. doi:10.1371/journal.pbio.0020073. PMC 368168. PMID 15024422. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=368168.
- ^ N. C. Seeman (2003). "DNA in a material world". Nature 421 (6921): 427–431. doi:10.1038/nature01406. PMID 12540916.
- ^ J. Chen & N. C. Seeman (1991). "Synthesis from DNA of a molecule with the connectivity of a cube" (w). Nature 350 (6319): 631–633. doi:10.1038/350631a0. PMID 2017259. http://www.palgrave-journals.com/doifinder/10.1038/350631a0
- ^ C. A. Mirkin, R. L. Letsinger, R. C. Mucic & J. J. Storhoff (1996). "A DNA-based method for rationally assembling nanoparticles into macroscopic materials". Nature 382 (6592): 607–609. doi:10.1038/382607a0. PMID 8757129.
- ^ H. Yan, S. H. Park, G. Finkelstein, J. H. Reif & T. H. Labean (2003). "DNA-Templated Self-Assembly of Protein Arrays and Highly Conductive Nanowires". Science 301 (5641): 1882–1884. doi:10.1126/science.1089389. PMID 14512621. http://www.sciencemag.org/cgi/content/abstract/301/5641/1882
- ^ J. S. Foster & J. E. Frommer (1988). "Imaging of liquid crystals using a tunnelling microscope". Nature 333 (6173): 542–545. Bibcode 1988Natur.333..542F. doi:10.1038/333542a0. http://www.nature.com/nature/journal/v333/n6173/abs/333542a0.html.
- ^ J.P. Rabe & S. Buchholz (1991). "Commensurability and Mobility in Two-Dimensional Molecular Patterns on Graphite". Science 353 (5018): 424–427. Bibcode 1991Sci...253..424R. doi:10.1126/science.253.5018.424. JSTOR 2878886.
External and further reading
- "Challenges and breakthroughs in recent research on self-assembly" Sci. Technol. Adv. Mater. 9 No 1(2008) 014109 (96 pages) free download
- G Kurth, Dirk (2008). "Metallo-supramolecular modules as a paradigm for materials science" (free-download review). Science and Technology of Advanced Materials 9: 014103. doi:10.1088/1468-6996/9/1/014103.
- Bureekaew, Sareeya; Shimomura, Satoru; Kitagawa, Susumu (2008). "Chemistry and application of flexible porous coordination polymers" (free-download review). Science and Technology of Advanced Materials 9: 014108. doi:10.1088/1468-6996/9/1/014108.
- H.E. Hoster, M. Roos, A. Breitruck, C. Meier, K. Tonigold, T. Waldmann, U. Ziener, K. Landfester, R.J. Behm, Structure Formation in Bis(terpyridine)Derivative Adlayers – Molecule-Substrate vs. Molecule-Molecule Interactions, Langmuir 23 (2007) 11570
- Molecular Self-Assembly papers
- Beyond molecules: Self-assembly of mesoscopic and macroscopic components
- Whitesides, G. M. & Grzyboski, B. (2002) Science 295, 2418-2421.
- Rothemund PWK, Papadakis N, Winfree E (2004). "Algorithmic Self-Assembly of DNA Sierpinski Triangles". PLoS Biol 2 (12): 12. doi:10.1371/journal.pbio.0020424. PMC 534809. PMID 15583715. http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0020424.
- C2 Wiki: Self Assembly from a computer programming perspective.
- Structure and Dynamics of Organic Nanostructures
- Metal organic coordination networks of oligopyridines and Cu on graphite
Nanotechnology (portal) Overview Nanomaterials Nanomedicine Molecular self-assembly Nanoelectronics Scanning probe microscopy Atomic force microscope · Scanning tunneling microscopeMolecular nanotechnology Categories:
Wikimedia Foundation. 2010.

