- Molecular recognition
-
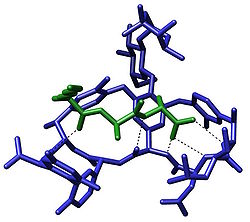 Crystal structure of a short peptide L-Lys-D-Ala-D-Ala (bacterial cell wall precursor) bound to the antibiotic vancomycin through hydrogen bonds[1]
Crystal structure of a short peptide L-Lys-D-Ala-D-Ala (bacterial cell wall precursor) bound to the antibiotic vancomycin through hydrogen bonds[1]
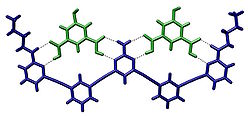 Crystal structure of two isophthalic acids bound to a host molecule through hydrogen bonds[2]
Crystal structure of two isophthalic acids bound to a host molecule through hydrogen bonds[2]
 Static recognition between a single guest and a single host binding site. In dynamic recognition binding the first guest at the first binding site induces a conformation change that affects the association constant of the second guest at the second binding site. In this case it is positive allosteric system.
Static recognition between a single guest and a single host binding site. In dynamic recognition binding the first guest at the first binding site induces a conformation change that affects the association constant of the second guest at the second binding site. In this case it is positive allosteric system.
The term molecular recognition refers to the specific interaction between two or more molecules through noncovalent bonding such as hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π-π interactions, electrostatic and/or electromagnetic[3] effects. The host and guest involved in molecular recognition exhibit molecular complementarity.[4][5]
Contents
Biological systems
Molecular recognition plays an important role in biological systems and is observed in between receptor-ligand, antigen-antibody, DNA-protein, sugar-lectin, RNA-ribosome, etc. An important example of molecular recognition is the antibiotic vancomycin that selectively binds with the peptides with terminal D-alanyl-D-alanine in bacterial cells through five hydrogen bonds. The vancomycin is lethal to the bacteria since once it has bound to these particular peptides they are unable to be used to construct the bacteria’s cell wall.
Supramolecular systems
Chemists have demonstrated that artificial supramolecular systems can be designed that exhibit molecular recognition. One of the earliest examples of such a system are crown ethers which are capable of selectively binding specific cations. However, a number of artificial systems have since been established.
Static vs. dynamic
Molecular recognition can be subdivided into static molecular recognition and dynamic molecular recognition. Static molecular recognition is likened to the interaction between a key and a keyhole; it is a 1:1 type complexation reaction between a host molecule and a guest molecule to form a host-guest complex. To achieve advanced static molecular recognition, it is necessary to make recognition sites that are specific for guest molecules.
In the case of dynamic molecular recognition the binding of the first guest to the first binding site of a host affects the association constant of a second guest with a second binding site.[6] In the case of positive allosteric systems the binding of the first guest increases the association constant of the second guest. While for negative allosteric systems the binding of the first guest decreases the association constant with the second. The dynamic nature of this type of molecular recognition is particularly important since it provides a mechanism to regulate binding in biological systems. Dynamic molecular recognition may enhance the ability to discriminate between several competing targets via the conformational proofreading mechanism. Dynamic molecular recognition is also being studied for application in highly functional chemical sensors and molecular devices.
See also
References
- ^ Knox, James R.; Pratt, R. F. (July 1990). "Different modes of vancomycin and D-alanyl-D-alanine peptidase binding to cell wall peptide and a possible role for the vancomycin resistance protein" (Free full text). Antimicrobial agents and chemotherapy 34 (7): 1342–7. PMC 175978. PMID 2386365. http://aac.asm.org/cgi/reprint/34/7/1342.
- ^ Bielawski, Christopher; Chen, Yuan-Shek; Zhang, Peng; Prest, Peggy-Jean; Moore, Jeffrey S. (1998). "A modular approach to constructing multi-site receptors for isophthalic acid" (Free full text). Chemical Communications (12): 1313–4. doi:10.1039/a707262g. http://www.rsc.org/delivery/_ArticleLinking/DisplayArticleForFree.cfm?doi=a707262g&JournalCode=CC.
- ^ Cosic, I (1994). "Macromolecular bioactivity: is it resonant interaction between macromolecules?—theory and applications". IEEE transactions on bio-medical engineering 41 (12): 1101–14. doi:10.1109/10.335859. PMID 7851912.
- ^ Lehn, Jean-Marie (1995). Supramolecular Chemistry. Weinheim: Wiley-VCH. ISBN 978-3-527-29312-4. OCLC 315928178.[page needed]
- ^ Gellman, Samuel H. (1997). "Introduction: Molecular Recognition". Chemical reviews 97 (5): 1231–1232. doi:10.1021/cr970328j. PMID 11851448.
- ^ Shinkai, Seiji; Ikeda, Masato; Sugasaki, Atsushi; Takeuchi, Masayuki (2001). "Positive allosteric systems designed on dynamic supramolecular scaffolds: toward switching and amplification of guest affinity and selectivity". Accounts of chemical research 34 (6): 494–503. doi:10.1021/ar000177y. PMID 11412086.
External links
- http://www.mdpi.org/ijms/sections/molecular-recognition.htm Special Issue on Molecular Recognition in the Int. J. Mol. Sci.
Categories:
Wikimedia Foundation. 2010.
