- Multicopper oxidase
-
Multicopper oxidase (type 1) 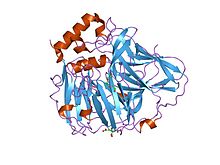
crystal structures of e. coli laccase cueo under different copper binding situations Identifiers Symbol Cu-oxidase Pfam PF00394 Pfam clan CL0026 InterPro IPR001117 PROSITE PDOC00076 SCOP 1aoz Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Multicopper oxidase (type 2) 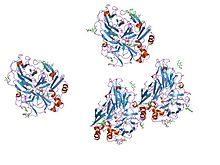
active laccase from trametes versicolor complexed with 2,5-xylidine Identifiers Symbol Cu-oxidase_2 Pfam PF07731 Pfam clan CL0026 InterPro IPR011706 SCOP 1aoz Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Multicopper oxidase (type 3) 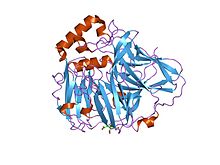
crystal structures of e. coli laccase cueo under different copper binding situations Identifiers Symbol Cu-oxidase_3 Pfam PF07732 Pfam clan CL0026 InterPro IPR011707 SCOP 1aoz Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary CMulti-copper polyphenol oxidoreductase laccase 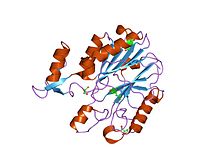
crystal structure of protein cc_0490 from caulobacter crescentus, pfam duf152 Identifiers Symbol Cu-oxidase_4 Pfam PF02578 InterPro IPR003730 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary In molecular biology, multicopper oxidases are enzymes which oxidise their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre; dioxygen binds to the trinuclear centre and, following the transfer of four electrons, is reduced to two molecules of water.[1] There are three spectroscopically different copper centres found in multicopper oxidases: type 1 (or blue), type 2 (or normal) and type 3 (or coupled binuclear).[2][3] Multicopper oxidases consist of 2, 3 or 6 of these homologous domains, which also share homology to the cupredoxins azurin and plastocyanin. Structurally, these domains consist of a cupredoxin-like fold, a beta-sandwich consisting of 7 strands in 2 beta-sheets, arranged in a Greek-key beta-barrel.[4] Multicopper oxidases include:
- Ceruloplasmin EC 1.16.3.1 (ferroxidase), a 6-domain enzyme found in the serum of mammals and birds that oxidizes different inorganic and organic substances; exhibits internal sequence homology that appears to have evolved from the triplication of a Cu-binding domain similar to that of laccase and ascorbate oxidase.
- Laccase EC 1.10.3.2 (urishiol oxidase), a 3-domain enzyme found in fungi and plants, which oxidizes different phenols and diamines. CueO is a laccase found in Escherichia coli that is involved in copper-resistance.[4]
- Nitrite reductase EC 1.7.2.1, a 2-domain enzyme containing type-1 and type-2 copper centres.[5][6]
In addition to the above enzymes there are a number of other proteins that are similar to the multi-copper oxidases in terms of structure and sequence, some of which have lost the ability to bind copper. These include: copper resistance protein A (copA) from a plasmid in Pseudomonas syringae; domain A of (non-copper binding) blood coagulation factors V (Fa V) and VIII (Fa VIII) [7]; yeast FET3 required for ferrous iron uptake [8]; yeast hypothetical protein YFL041w; and the fission yeast homologue SpAC1F7.08.
References
- ^ Bento I, Martins LO, Gato Lopes G, Arménia Carrondo M, Lindley PF (November 2005). "Dioxygen reduction by multi-copper oxidases; a structural perspective". Dalton Trans (21): 3507–13. doi:10.1039/b504806k. PMID 16234932.
- ^ Messerschmidt A, Huber R (January 1990). "The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships". Eur. J. Biochem. 187 (2): 341–52. doi:10.1111/j.1432-1033.1990.tb15311.x. PMID 2404764.
- ^ Ouzounis C, Sander C (February 1991). "A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins". FEBS Lett. 279 (1): 73–8. doi:10.1016/0014-5793(91)80254-Z. PMID 1995346.
- ^ a b Roberts SA, Weichsel A, Grass G, Thakali K, Hazzard JT, Tollin G, Rensing C, Montfort WR (March 2002). "Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli". Proc. Natl. Acad. Sci. U.S.A. 99 (5): 2766–71. doi:10.1073/pnas.052710499. PMC 122422. PMID 11867755. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=122422.
- ^ Nakamura K, Kawabata T, Yura K, Go N (October 2003). "Novel types of two-domain multi-copper oxidases: possible missing links in the evolution". FEBS Lett. 553 (3): 239–44. doi:10.1016/S0014-5793(03)01000-7. PMID 14572631.
- ^ Suzuki S, Kataoka K, Yamaguchi K (October 2000). "Metal coordination and mechanism of multicopper nitrite reductase". Acc. Chem. Res. 33 (10): 728–35. PMID 11041837.
- ^ Mann KG, Jenny RJ, Krishnaswamy S (1988). "Cofactor proteins in the assembly and expression of blood clotting enzyme complexes". Annu. Rev. Biochem. 57: 915–56. doi:10.1146/annurev.bi.57.070188.004411. PMID 3052293.
- ^ Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J (January 1994). "The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake". Cell 76 (2): 403–10. doi:10.1016/0092-8674(94)90346-8. PMID 8293473.
This article includes text from the public domain Pfam and InterPro IPR001117
This article includes text from the public domain Pfam and InterPro IPR011706
This article includes text from the public domain Pfam and InterPro IPR011707
This article includes text from the public domain Pfam and InterPro IPR003730
Categories:- Protein domains
Wikimedia Foundation. 2010.
