- Diguanylate cyclase
-
diguanylate cyclase 
Crystal structure of diguanylate cyclase PleD in complex with c-di-GMP from Caulobacter crescentus; rendering based on PDB 2WB4 Identifiers EC number 2.7.7.65 CAS number 146316-82-7 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles In enzymology, diguanylate cyclase, also known as diguanylate kinase (EC 2.7.7.65), is an enzyme that catalyzes the chemical reaction:
2 Guanosine triphosphate ↔ 2 diphosphate + cyclic di-3',5'-guanylate
The substrates of diguanylate cyclases (DGCs) are two molecules of guanosine triphosphate (GTP) and the products are two molecules of diphosphate and one molecule of cyclic di-3’,5’-guanylate (cyclic di-GMP).
Degradation of cyclic di-GMP to guanosine monophosphate (GMP) is catalyzed by a phosphodiesterase (PDE).
Contents
Structure
Diguanylate cyclases are characterized by the conserved amino acid sequence motifs “GGDEF” (Gly-Gly-Asp-Glu-Phe) or “GGEEF” (Gly-Gly-Glu-Glu-Phe), which constitute the domain of the DGC active site.[1] These domains are often found coupled to other signaling domains within multidomain proteins. Oftentimes, GGDEF domains with DGC activity are found in the same proteins as c-di-GMP-specific phosphodiesterase (PDE) EAL (Glu-Ala-Leu) domains.[2][3]
DGC is thought to only be active as a dimer consisting of two subunits, both with GGDEF domains.[4] The active (or catalytic) site is located at the interface between the two subunits, each binding one molecule of GTP. (See Activation mechanism and Regulation section for more information)
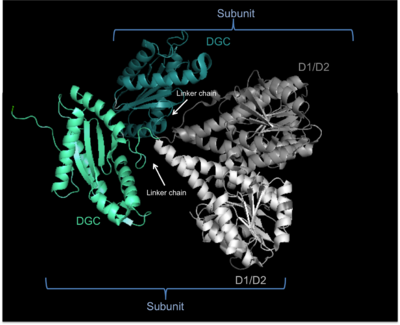
Weak sequence similarity and pronounced secondary structure similarity between GGDEF domains and the catalytic domains of adenylate cyclases (AC) have led to the hypothesis that DGCs and ACs share a similar fold.[5] This was verified with the resolution of the crystal structure of the DGC PleD from Caulobacter crescentus in complex with c-di-GMP.[4] As shown in the figure, active PleD, shown as a dimer, is composed of the catalytic DCG domain (labeled DGC) and two CheY-like receiver domains (labeled D1/D2). The DGC domain of each subunit is linked to the two CheY-like domains by a flexible peptide linkage chain.[4] The DCG domain closely resembles the domain of the AC catalytic core which consists of a five-stranded β-sheet surrounded by helices.
As of mid 2011, 11 crystal structures of confirmed or putative DGCs have been solved, with PDB accession codes PDB 3N53, PDB 3N3T, PDB 3MTK, PDB 2WB4, PDB 3KZP, PDB 3HVA, PDB 3I5A, PDB 3IGN, PDB 3HVW, PDB 3H9W, and PDB 2R60.
Biological function
Diguanylate cyclase participate in the formation of the ubiquitous second messenger, cyclic-di-GMP, involved in bacterial biofilm formation and persistence. The GGDEF domain was first identified in the regulatory protein, PleD of the bacterium Caulobacter crescentus.[6] It was later noted that numerous bacterial genomes encoded multiple proteins with a GGDEF domain.[7] Pseudomonas aeruginosa PAO1 has 33 proteins with GGDEF domains, Escherichia coli K-12 has 19, and Vibrio cholerae O1 has 41.[8] In the cell cycle of Caulobacter crescentus, DGC PleD is known to control pole morphogenesis.[9] In Pseudomonas fluorescens DGC WspR activity is hypothesized to be partially responsible for the wrinkly spreader (WS) phenotype.[10] In Pseudomonas aeruginosa, WspR has also been known to control autoaggregation.[8]
Role of DGC in C. crescentus cell cycle
During the cell cycle of "C. cresentus", proteins with GGDEF and EAL domains are separated towards the two distinct poles. The active form of diguanylate cyclase PleD localizes to the stalked pole of differentiating C. crescentus cells.[11] It has been suggested that the function of PleD is two-fold. Pled is responsible for turning off flagellum rotations and inhibiting motility before genome replication begins and also for regenerating motility after differentiation has completed.[12]
 Activation Mechanism and Regulation through Product Inhibition of the DGC PleD from C. crescentus [4]
Activation Mechanism and Regulation through Product Inhibition of the DGC PleD from C. crescentus [4]
Activation Mechanism and Regulation
The crystal structure of the C. crescentus diguanylate cyclase, PleD, contains three domains; a GGDEF domain with diguanylate cyclase activity and two CheY-like receiver domains (D1/D2). As seen in the figure, the active form of PleD is a dimer which forms by phosphorylation of the first receiver domain (D1).[4] Phosphorylation of the receiver domain increases the dimerization affinity by approximately 10-fold over non-phosphorylated domains.[2][13]
Inhibition of DGC activity is thought to be allosteric and non-competitive.[4][14] Cyclic di-GMP binds to interface between the DGC and D2 domains stabilizing the open structure and preventing catalysis.[15] Strong product inhibition has been observed with a Ki of 0.5 μM.[4]
Though the exact catalytic mechanism has not been resolved, it is hypothesized that the dimerized structure of PleD facilitates interaction of the two GTP molecules within the DGC active site for cyclization. A proposed mechanism by Chan et al. indicates that the 3'-OH group of the GTP is deprotonated by a glutamic acid residue (E370) to allow for intermolecular nucleophilic attack of the α-phosphate. The pentachoordinated transition state created through this nucleophilic attack is possibly stabilized by a Lysine residue (K332).
References
- ^ Ausmees, N.,, Mayer, R., Weinhouse, H., Volman, G., Amikam, D., Benziman, M., Lindberg, M. (October 2001). "Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity". FEMS Microbiology Letters 204 (1): 163–167. doi:10.1111/j.1574-6968.2001.tb10880.x. PMID 11682196.
- ^ a b Stock, A.M. (August 2007). "Diguanylate Cyclase Activation: It Takes Two". Structure 15 (8): 887–888. doi:10.1016./j.str.2007.07.003. PMID 17697992.
- ^ Ryjenkov, D., Tarutina, M., Moskvin, O.V., Gomelsky, M. (March 2005). "Cyclic Diguanylate Is a Ubiquitous Signaling Molecule in Bacteria Inights into Biochemistry of the GGDEF Protein Domain". Journal of Bacteriology 187 (5): 1792–1798. doi:10.1128/JB.187.5.1792-1798.2005. PMC 1064016. PMID 15716451. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1064016.
- ^ a b c d e f g Chan, C., Paul, R., Samoray, D., Amiot, N.C., Giese, B., Jenal, U., Schirmer, T. (December 2004). "Structural basis of activity and allosteric control of diguanylate cyclase". PNAS 101 (49): 17084–17089. doi:10.1073/pnas.0406134101. PMC 535365. PMID 15569936. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=535365.
- ^ Pei, J., Grishin, N. (February 2001). "GGDEF domain is homologous to adenylyl cyclase". Proteins 42 (2): 210–216. doi:10.1002/1097-0134(20010201)42:2<210::AID-PROT80>3.0.CO;2-8. PMID 11119645.
- ^ Hecht, G.B., Newton, A. (1995). "Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus". Journal of Bacteriology 177 (21): 6223–6229. PMC 177463. PMID 7592388. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=177463.
- ^ Galperin, M.Y., Nikolskaya, A.N., Koonin, E.V. (2001). "Novel domains of the prokaryotic two-component signal transduction systems". FEMS Microbial Letters 203 (1): 11–21. doi:10.1016/S0378-1097(01)00326-3. PMID 11557134.
- ^ a b D'Argenio, D.A., Miller, S.I. (2004). "Cyclic di-GMP as a bacterial second messenger". Microbiology 150 (Pt 8): 2797–2502. doi:10.1099/mic.0.27099-0. PMID 15289546.
- ^ Aldridge P, Paul R, Goymer P, Rainey P, Jenal U (March 2003). "Role of the GGDEF-regulated PleD in polar development of Caulobacter crescentus". Molecular Microbiology 47 (6): 1695–1708. doi:10.1046/j.1365-2958.2003.03401.x. PMID 12622822.
- ^ Malone J.G., Williams R., Christen M, Jenal U, Spiers A.J., Rainey P.B., (December 2006). "The structure-function relationship of WspR, a Psuedomonas fluorescens response regulator with a GGDEF output domain". Microbiology 153 (Pt 4): 980–153. doi:10.1099/mic.0.2006/002824-0. PMID 17379708.
- ^ Paul, R., Weiser, S., Amiot, N.C., Chan, C., Schirmer, T., Giese, B., Jenal, U. (2004). "Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain". Genes & Development 18 (6): 715–727. doi:10.1101/gad.289504. PMC 387245. PMID 15075296. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=387245.
- ^ Skerker, J.M., Laub, M.T. (April 2004). "Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus". Nature Reviews: Microbiology 18 (4): 335–337. doi:10.1038/nrmicro864. PMID 15031731.
- ^ Wassmann, P., Chan, C., Paul, R., Beck, A., Heerklotz, H., Jenal, U. (August 2007). "Structure of BeF3- - Modified Response Regulator PleD: Implications for Diguanylate Cyclase Activation, Catalysis, and Feedback Inhibition". Structure 15 (8): 915–927. doi:10.1016/j.str.2007.06.016. PMID 17697997.
- ^ Paul, R,. Abel, S., Wassmann, P., Beck, A., Heerklotz, H., Jenal, U. (October 2007). "Activation of the Diguanylate Cyclase PleD by Phosphorylation-mediated Dimerization". Journal of Biological Chemistry 282 (40): 29170–29177. doi:10.1074/jbc.M704702200. PMID 17640875.
- ^ Christen, B., Christen, M., Paul, R., Schmid, F., Folcher, M., Jenoe, P., Meuwly, M., (October 2006). "Allosteric Control of Cyclic di-GMP Signaling". Journal of Biological Chemistry 281 (42): 32015–32024. doi:10.1074/jbc.M603589200. PMID 16923812.
Further reading
- Schirmer, T and Urs Jenal (2009). "Structural and mechanistic determinants of d-di-GMP signaling". Nature 7 (10): 724–735. doi:10.1038/nrmicro2203. PMID 19756011.
- Jenal, U. and Jacob Malone (2006). "Mechanisms of Cyclic-di-GMP Signaling in Bacteria". Review 40: 385–407. doi:10.1146/annurev.genet.40.110405.090423. PMID 16895465.
- Hengge, R. (2009). "Principles of c-di-GMP signalling in bacteria". Review 7 (4): 263–273. doi:10.1038/nrmicro2109. PMID 19287449.
Categories:- Enzymes
Wikimedia Foundation. 2010.