- Malassezia
-
Malassezia 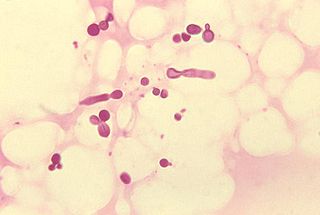
Malassezia furfur in skin scale from a patient with tinea versicolor Scientific classification Kingdom: Fungi Division: Basidiomycota Subdivision: Ustilaginomycotina Class: Exobasidiomycetes Order: Malasseziales Family: Malasseziaceae Genus: Malassezia Species See text.
Malassezia (formerly known as Pityrosporum) is a genus of fungi. Malassezia is naturally found on the skin surfaces of many animals, including humans. In occasional opportunistic infections, some species can cause hypopigmentation on the trunk and other locations in humans. Allergy tests for this fungus are available.
Contents
Nomenclature
Due to progressive changes in their nomenclature, some confusion exists about the naming and classification of Malassezia yeast species. Work on these yeasts has been complicated because they are extremely difficult to propagate in laboratory culture.
Malassezia were originally identified by the French scientist Louis-Charles Malassez in the late 19th century. Raymond Sabouraud identified a dandruff-causing organism in 1904 and called it "Pityrosporum malassez", honoring Malassez, but at the species level as opposed to the genus level. When it was determined that the organisms were the same, the term "Malassezia" was judged to possess priority.[1]
In the mid-20th century, it was reclassified into two species:
- Pityrosporum (Malassezia) ovale, which is lipid-dependent and found only on humans. P. ovale was later divided into two species, P. ovale and P. orbiculare, but current sources consider these terms to refer to a single species of fungus, with M. furfur the preferred name.[2]
- Pityrosporum (Malassezia) pachydermatis, which is lipophilic but not lipid-dependent, is found on the skin of most animals.
In the mid-1990s, scientists at the Pasteur Institute in Paris, France, discovered additional species.[3]
Currently there are 10 recognized species:
- M. furfur
- M. pachydermatis[4]
- M. globosa[5]
- M. restricta[6]
- M. slooffiae[7]
- M. sympodialis[8]
- M. nana[9]
- M. yamatoensis[10]
- M. dermatis[11]
- M. obtusa
Role in human diseases
Recently, identification of Malassezia on skin has been aided by the application of molecular or DNA-based techniques very similar to those used by forensic scientists to identify criminal suspects. These investigations show that the Malassezia species causing most skin disease in humans, including the most common cause of dandruff and seborrhoeic dermatitis, is M. globosa (though M. restricta is also involved).[5] The skin rash of tinea versicolor (pityriasis versicolor) is also due to infection by this fungus.
As the fungus requires fat to grow, it is most common in areas with many sebaceous glands: on the scalp,[12] face, and upper part of the body. When the fungus grows too rapidly, the natural renewal of cells is disturbed and dandruff appears with itching (a similar process may also occur with other fungi or bacteria).
A project in 2007 has sequenced the genome of dandruff-causing Malassezia globosa and found it to have 4,285 genes.[13] M. globosa uses eight different types of lipase, along with three phospholipases, to break down the oils on the scalp. Any of these 11 proteins would be a suitable target for dandruff medications.
M. globosa has been predicted to have the ability to reproduce sexually,[13][14] but this has not been observed.
Numbers
The number of specimens of M. globosa on a human head can be up to ten million.
Treatment of symptomatic scalp infections
Further information: Seborrhoeic_dermatitis#TreatmentsSymptomatic scalp infections are often treated with selenium disulfide[15] or ketoconazole containing shampoos. Other treatments include ciclopirox olamine, coal tar, zinc pyrithione (ZPT), miconazole, or tea tree oil medicated shampoos. Used occasionally and diluted with water, Hydrogen peroxide is also used to manage symptoms of itching. However, with its oxidative capability in reaction with catalase, scarring occurs with this treatment.
References
- ^ Inamadar AC, Palit A (2003). "The genus Malassezia and human disease". Indian J Dermatol Venereol Leprol 69 (4): 265–70. PMID 17642908. http://www.ijdvl.com/article.asp?issn=0378-6323;year=2003;volume=69;issue=4;spage=265;epage=270;aulast=Inamadar.
- ^ Freedberg et al, ed (2003). Fitzpatrick's Dermatology in General Medicine. (6th ed.). McGraw-Hill. pp. 1187. ISBN 0-07-138067-1.
- ^ Guého E, Midgley G, Guillot J (May 1996). "The genus Malassezia with description of four new species". Antonie Van Leeuwenhoek 69 (4): 337–55. doi:10.1007/BF00399623. PMID 8836432.
- ^ Coutinho SD, Paula CR (June 1998). "Biotyping of Malassezia pachydermatis strains using the killer system". Rev Iberoam Micol 15 (2): 85–7. PMID 17655416. http://www.reviberoammicol.com/pubmed_linkout.php?15p85.
- ^ a b DeAngelis YM, Saunders CW, Johnstone KR, et al. (September 2007). "Isolation and expression of a Malassezia globosa lipase gene, LIP1". J. Invest. Dermatol. 127 (9): 2138–46. doi:10.1038/sj.jid.5700844. PMID 17460728.
- ^ Sugita T, Tajima M, Amaya M, Tsuboi R, Nishikawa A (2004). "Genotype analysis of Malassezia restricta as the major cutaneous flora in patients with atopic dermatitis and healthy subjects" (– Scholar search). Microbiol. Immunol. 48 (10): 755–9. PMID 15502408. http://joi.jlc.jst.go.jp/JST.JSTAGE/mandi/48.755?from=PubMed.[dead link]
- ^ Uzal FA, Paulson D, Eigenheer AL, Walker RL (October 2007). "Malassezia slooffiae-associated dermatitis in a goat". Vet. Dermatol. 18 (5): 348–52. doi:10.1111/j.1365-3164.2007.00606.x. PMID 17845623. http://www3.interscience.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=0959-4493&date=2007&volume=18&issue=5&spage=348.
- ^ Niamba P, Weill FX, Sarlangue J, Labrèze C, Couprie B, Taïeh A (August 1998). "Is common neonatal cephalic pustulosis (neonatal acne) triggered by Malassezia sympodialis?". Arch Dermatol 134 (8): 995–8. doi:10.1001/archderm.134.8.995. PMID 9722730. http://archderm.ama-assn.org/cgi/pmidlookup?view=long&pmid=9722730.
- ^ Hirai A, Kano R, Makimura K, et al. (March 2004). "Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals". Int. J. Syst. Evol. Microbiol. 54 (Pt 2): 623–7. doi:10.1099/ijs.0.02776-0. PMID 15023986. http://ijs.sgmjournals.org/cgi/pmidlookup?view=long&pmid=15023986.
- ^ Sugita T, Tajima M, Takashima M, et al. (2004). "A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects" (– Scholar search). Microbiol. Immunol. 48 (8): 579–83. PMID 15322337. http://joi.jlc.jst.go.jp/JST.JSTAGE/mandi/48.579?from=PubMed.[dead link]
- ^ Sugita T, Takashima M, Shinoda T, et al. (April 2002). "New Yeast Species, Malassezia dermatis, Isolated from Patients with Atopic Dermatitis". J. Clin. Microbiol. 40 (4): 1363–7. doi:10.1128/JCM.40.4.1363-1367.2002. PMC 140359. PMID 11923357. http://jcm.asm.org/cgi/pmidlookup?view=long&pmid=11923357.
- ^ "Genetic code of dandruff cracked". BBC News. 6 November 2007. http://news.bbc.co.uk/2/hi/health/7080434.stm. Retrieved 2008-12-10.
- ^ a b Xu J, Saunders CW, Hu P, et al. (November 2007). "Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens". Proc. Natl. Acad. Sci. U.S.A. 104 (47): 18730–5. doi:10.1073/pnas.0706756104. PMC 2141845. PMID 18000048. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=18000048.
- ^ Guillot J, Hadina S, Guého E (June 2008). "The genus Malassezia: old facts and new concepts". Parassitologia 50 (1–2): 77–9. PMID 18693563.
- ^ DermNet treatments/selenium Accessed Dec 24, 2007.
External links
- M Shams Ghahfarokhi, M. Razzaghi Abyaneh (March 2004). "Rapid Identification of Malassezia furfur from other Malassezia Species: A Major Causative Agent of Pityriasis Versicolor" (PDF). IJMS 29 (1). http://ijms.sums.ac.ir/files/PDFfiles/29_1_08-Shams.pdf.
- Noble SL, Forbes RC, Stamm PL (July 1998). "Diagnosis and management of common tinea infections". Am Fam Physician 58 (1): 163–74, 177–8. PMID 9672436. http://www.aafp.org/afp/980700ap/noble.html.
Categories:- Agaricomycotina
- Parasitic fungi
- Yeasts
Wikimedia Foundation. 2010.
