- UCSF Chimera
-
UCSF Chimera 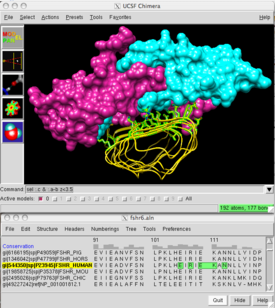
Chimera main window (FSH and receptor, 1xwd) and sequence window (alignment of FSH receptors from different species).Developer(s) Resource for Biocomputing, Visualization, and Informatics (RBVI), UCSF Stable release 1.5.3 / May 24, 2011 Operating system Windows, Mac OS X, Linux Type Molecular modelling License free for noncommercial use Website www.cgl.ucsf.edu/chimera UCSF Chimera (or simply Chimera) is an extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories, and conformational ensembles. High-quality images and movies can be created. Chimera includes complete documentation and can be downloaded free of charge for noncommercial use.
Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics (RBVI) at the University of California, San Francisco. Development is funded by the NIH National Center for Research Resources (grant P41-RR001081).
Contents
General Structure Analysis
- automatic atom type identification
- hydrogen addition and partial charge assignment
- high-quality hydrogen bond, contact, and clash detection
- distance and angle measurements, rotatable bonds, including Ramachandran plot visualization
- amino acid rotamer libraries (backbone-dependent and -independent)
- analysis of metal coordination geometry
- molecular dynamics trajectory playback (many formats)
- clustering of conformational ensembles
- morphing between conformations of a protein or even different proteins
- calculation of theoretical SAXS profiles
- display of attributes (B-factor, hydrophobicity, etc.) with colors, radii, "worms"
- easy creation of new attributes with simple text file inputs
- rich set of commands, powerful specification syntax
- many formats read, PDB and Mol2 written
- web fetch from Protein Data Bank, EDS (density maps), EMDB (density maps), ModBase (comparative models), CASTp (protein pocket measurements), Pub3D (small molecule structures), VIPERdb (icosahedral virus capsids), others
Presentation Images and Movies
- images can be saved at arbitrarily high resolution
- adjustable background color, lighting, depth-cueing, silhouette edges
- stick, ball-and-stick, sphere, ribbon, and surface displays
- ellipsoids to show anisotropic B-factors
- nonmolecular geometric objects
- labeling with arbitrary text, symbols, arrows, color keys
- isosurfaces and transparent renderings of volume data (see below)
- different structures can be clipped differently and at any angle
- optional raytracing with bundled POV-Ray
- scene export to X3D and other formats
- simple graphical interface for creating movies interactively
- alternatively, movie content and recording can be scripted
- movie recording is integrated with morphing and MD trajectory playback
Volume Data Tools
- many formats of volume data maps (electron density, electrostatic potential, others) read, several written
- interactive threshold adjustment, multiple isosurfaces (mesh or solid), transparent renderings
- fitting of atomic coordinates to maps and maps to maps
- density maps can be created from atomic coordinates
- markers can be placed in maps and connected with smooth paths
- display of single data planes, optionally as topographic maps
- volume data time series playback and morphing
- many tools for segmenting and editing maps
- Gaussian smoothing, Fourier transform
- surface area and surface-enclosed volume measurements
Sequence-Structure Tools
- many sequence alignment formats read, written
- sequence alignments can be created, edited
- structures automatically associate with sequences, with some mismatch tolerance
- sequence-structure crosstalk: highlighting in one highlights the other
- multiple methods for calculating conservation and displaying values on associated structures
- user-defined headers (lines above the sequences) including histograms and colored symbols
- trees in Newick format read/displayed
- protein BLAST search via Web service
- interface to MODELLER for homology modeling
- structure superposition using the associated sequence alignment
- in the absence of a pre-existing alignment, structure superposition using both residue type and secondary structure information
- generation of structure-based sequence alignments from superpositions of two or more proteins
See also
- List of molecular graphics systems
- Molecular modelling
- Molecular graphics
- Molecular dynamics
- Software for molecular mechanics modeling
External links
Categories:- Molecular modelling software
- Bioinformatics software
- Science software
- Freeware
Wikimedia Foundation. 2010.
