- Maitotoxin
-
Not to be confused with mitotoxin.
Maitotoxin 
Identifiers CAS number 59392-53-9 
Properties Molecular formula C164H256O68S2Na2 Molar mass 3422 g/mol  (verify) (what is:
(verify) (what is:  /
/ ?)
?)
Except where noted otherwise, data are given for materials in their standard state (at 25 °C, 100 kPa)Infobox references Maitotoxin (or MTX) is an extremely potent toxin produced by Gambierdiscus toxicus, a dinoflagellate species. Maitotoxin is so potent that it has been demonstrated that an intraperitoneal injection of 0.13 µg/kg was lethal in mice.[1] Maitotoxin was named from the ciguateric fish Ctenochaetus striatus—called "maito" in Tahiti—from which maitotoxin was isolated for the first time. It was later shown that maitotoxin is actually produced by Gambierdiscus toxicus.
Contents
Mechanism of toxicity
Maitotoxin activates Ca2+ permeable, non-selective cation channels, leading to an increase in levels of cytosolic Ca2+ ions. It is thought that maitotoxin leads to the formation of pores on these ion channels. Ultimately, a cell death cascade is activated, resulting in membrane blebbing and eventually cell lysis.[2] Maitotoxin is known to activate cytosolic calcium-activated proteases calpain-1 and calpain-2, contributing to necrosis.[3] The toxicity of maitotoxin to mice is the highest for nonprotein toxins: the LD50 is 50 ng/kg.
Molecular Structure
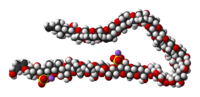
The molecule itself is a system of 32 fused rings. It is notable because it is one of the largest and most complex non-protein, non-polysaccharide molecules produced by an organism. Maitotoxin includes 32 ether rings, 22 methyls, 28 hydroxyls, and 2 sulfuric acid esters and has an amphipathic structure.[4][5][6] Its structure was established through analysis using nuclear magnetic resonance at Tohoku University, Harvard and the University of Tokyo in combination with mass spectrometry, and synthetic chemical methods. However, Gallimore and Spencer recently questioned the structure of maitotoxin at a single ring-junction (the J-K junction), based purely on biosynthetic considerations and their general model for marine polyether biogenesis.[7] Nicolaou and Frederick on the other hand argue that despite this biosynthetic argument, the originally proposed structure could still be correct. [8] The controversy has yet to be resolved.
Biosynthesis
The core synthetic route is via a polyketide synthase pathway.[7]
References
- ^ Yokoyama, A et al. (1988). "Some Chemical Properties of Maitotoxin, a Putative Calcium Channel Agonist Isolated from a Marine Dinoflagellate". J. Biochem 104 (2): 184–187. PMID 3182760.
- ^ Estacion, M and Schilling, WP (2001). "Maitotoxin-induced membrane blebbing and cell death in bovine aortic endothelial cells". BMC Physiology 1: 2. doi:10.1186/1472-6793-1-2.
- ^ Wang, K. et al. (1996). "Maitotoxin induces calpain activation in SH-SY5Y neuroblastoma cells and cerebrocortical cultures". Arch. Biochem. Biophys. 331 (2): 208–214. doi:10.1006/abbi.1996.0300. PMID 8660700.
- ^ Murata, M et al. (1994). "Structure and partial stereochemical assignments for maitotoxin, the most toxic and largest natural non-biopolymer". J. Am. Chem. Soc. 116 (16): 7098–7107. doi:10.1021/ja00095a013.
- ^ Sasaki, M et al. (1996). "The complete structure of maitotoxin, I; Configuration of the C1-C14 side chain". Angew. Chem. Int. Ed. Engl. 35 (15): 1672–1675. doi:10.1002/anie.199616721.
- ^ Kishi, Y (1998). "Complete structure of maitotoxin". Pure & Appl. Chem. 70 (2): 339–344. doi:10.1351/pac199870020339.
- ^ a b Gallimore AR, Spencer JB (2006). "Stereochemical Uniformity in Marine Polyether Ladders—Implications for the Biosynthesis and Structure of Maitotoxin". Angew. Chem. Int. Ed. Engl. 45 (27): 4406–4413. doi:10.1002/anie.200504284. PMID 16767782.
- ^ Nicolaou KC, Frederick MO (2007). "On the structure of maitotoxin". Angew. Chem. Int. Ed. Engl. 46 (28): 5278–82. doi:10.1002/anie.200604656. PMID 17469088.
Further reading
- Jones, Maitland (2004). Organic Chemistry, Third Edition. W. W. Norton & Company. ISBN 978-0393924084.
Categories:- Oxygen heterocycles
- Neurotoxins
Wikimedia Foundation. 2010.
