- Oxalyl-CoA decarboxylase
-
oxalyl-CoA decarboxylase 
Identifiers EC number 4.1.1.8 CAS number 9024-96-8 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles In enzymology, an oxalyl-CoA decarboxylase (OXC) (EC 4.1.1.8) is an enzyme primarily produced by the gastrointestinal bacterium Oxalobacter formigenes that catalyzes the chemical reaction
- oxalyl-CoA
 formyl-CoA + CO2
formyl-CoA + CO2
OXC belongs to the family of lyases, specifically the carboxy-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is oxalyl-CoA carboxy-lyase (formyl-CoA-forming). Other names in common use include oxalyl coenzyme A decarboxylase, and oxalyl-CoA carboxy-lyase. This enzyme participates in glyoxylate and dicarboxylate metabolism. It employs one cofactor, thiamin diphosphate (TPP), and plays a key role in catabolism of oxalate, a highly toxic compound that is a product of the oxidation of carbohydrates in many bacteria and plants[1]. Oxalyl-CoA decarboxylase is extremely important for the elimination of ingested oxalates found in human foodstuffs like coffee, tea, and chocolate,[2] and the ingestion of such foods in the absence of Oxalobacter formigenes in the gut can result in kidney disease or even death as a result of oxalate poisoning.[3]
Contents
Evolution
Oxalyl-CoA decarboxylase is hypothesized to be evolutionarily related to acetolactate synthase, a TPP-dependent enzyme responsible for the biosynthesis of branched chain amino acids in certain organisms.[4] Sequence alignments between the two enzymes support this claim, as do the presence of vestigial FAD-binding pockets that play no role in either enzyme’s catalytic activity.[5] The binding of FAD at this site in acetolactate synthase and the binding of ADP at a cognate site in OXC are thought to play roles in the stabilization of the tertiary structures of the proteins.[6] No FAD binding is observed in oxalyl-CoA decarboxylase,[7] but an excess of coenzyme A in the crystal structure has led to the hypothesis that the binding site was co-opted during OXC evolution to bind the CoA moiety of its substrate.[8] Despite their similarities, only oxalyl-CoA decarboxylase is necessary for the formation of ATP in Oxalobacter formigenes, and exogenous ADP has been been demonstrated to increase the decarboxylase activity of OXC, but not acetolactate synthase.[9][10]
Reaction Mechanism
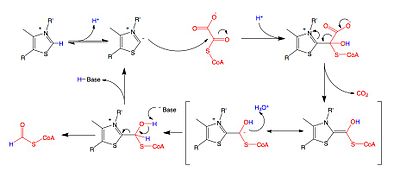
A key feature of the cofactor TPP is the relatively acidic proton bound to the carbon atom between the nitrogen and sulfur in the thiazole ring, which has a pKa near 10.[11] This carbon center ionizes to form a carbanion, which adds to the carbonyl group of oxalyl-CoA. This addition is followed by the decarboxylation of oxalyl-CoA, and then the oxidation and removal of formyl-CoA to regenerate the carbanion form of TPP. While the reaction mechanism is shared with other TPP-dependent enzymes, the residues found in the active site of OXC are unique, which has raised questions about whether TDP must be deprotonated by a basic amino acid at a second site away from the carbanion-forming site to activate the cofactor.[12]
Structure
 Two colorizations of the dimeric substructure of the enzyme. Left side distinguishes the enzyme's secondary structures and right side distinguishes the two monomers. Derived from 2JI6
Two colorizations of the dimeric substructure of the enzyme. Left side distinguishes the enzyme's secondary structures and right side distinguishes the two monomers. Derived from 2JI6
Oxalyl-CoA decarboxylase is tetrameric, and each monomer consists of three α/β-type domains.[13] The thiamine diphosphate-binding site rests on the subunit-subunit interface between two of the domains, which is commonly seen in its class of enzymes. Oxalyl-CoA decarboxylase is structurally homologous to acetolactate synthase found in plants and other microorganisms, but OXC binds ADP in a region that is similar to the FAD-binding site in acetolactate synthase.[14][15]
As of late 2007, 6 structures have been solved for this class of enzymes, with PDB accession codes 2C31, 2JI6, 2JI7, 2JI8, 2JI9, and 2JIB.
References
- ^ Baetz, A.L. and Allison, M.J. “Purification and characterization of formyl-coenzyme A transferase from Oxalobacter formigenes.” J. Bacteriol 172, 7 (1990).
- ^ Gasinka, A. and Gajewska, D. “Tea and Coffee as the Main Source of Oxalate in Diets of Patients with Kidney Oxalate Stones.” Roczn. Pzh 58, 1 (2007).
- ^ Turoni, S. et al. “Oxalate-degrading activity in Bifidobacterium animalis subsp. lactis: impact of acidic conditions on the transcriptional levels of the oxalyl coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes.” Appl. Environ. Microbiol. 76, 10 (2010).
- ^ Dailey, F.E and Cronan, J.E. “Acetohydroxy acid synthase I, a required enzyme for isoleucine and valine biosynthesis in Escherichia coli K-12 during growth on acetate as the sole carbon source." J. Bacteriol 165, 2 (1986).
- ^ Chipman, D. et al. “Biosynthesis of 2-aceto-2-hydroxy acids: acetolactate synthases and acetohydroxyacid synthases.” Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology 1385, 2 (1998).
- ^ Signh, B.K. and Schmitt, G.K. “Flavin adenine dinucleotide causes oligomerization of acetohydroxyacid synthase from black Mexican sweet corn cells.” FEBS Letters 258, 1 (1989).”
- ^ Svedruzic, D. et al. “The enzymes of oxalate metabolism: unexpected structures and mechanisms.” Archives of Biochemistry and Biophysics 433, 1 (2005).
- ^ Berthold, C.L. et al. “Crystallographic Snapshots of Oxalyl-CoA Decarboxylase Give Insights into Catalysis by Nonoxidative ThDP-Dependent Decarboxylases.” Structure 15, 7 (2007).
- ^ Maestri, O. and Joset, F. “Regulation by external pH and stationary growth phase of the acetolactate synthase from Synechocystis PCC6803.” Mol.Microbiol. 37, 4 (2000).
- ^ Whitlow, K.J. and Polgase, W.J. “Regulation of acetohydroxy acid synthase in streptomycin-dependent Escherichia coli.” J. Bacteriol 121, 1 (1970).
- ^ Berg, J.M, Tymoczko J.L., and Stryer, L. Biochemistry: 6th Edition. NY: W.H. Freeman and Company. Page 479.
- ^ Berthold C.L. et al. “Structural basis for activation of the thiamin diphosphate-dependent enzyme oxalyl-CoA decarboxylase by adenosine diphosphate.” J. Biol. Chem. 280, 50 (2005).
- ^ Werther, T. et al. “New insights into structure–function relationships of oxalyl CoA decarboxylase from Escherichia coli.” FEBS Journal 277, 12 (2010).
- ^ Dugglebay, R.J. and Pang, S.S. “Acetohydroxyacid Synthase.” Journal of Biochemistry and Molecular Biology 33, 1 (2000).
- ^ Azcarate-Peril, M.A. et al. “Transcriptional and Functional Analysis of Oxalyl-Coenzyme A (CoA) Decarboxylase and Formyl-CoA Transferase Genes fromLactobacillus acidophilus.” Appl. Environ. Microbiol. 72, 3 (2006).
Categories:- EC 4.1.1
- Thiamine enzymes
- Enzymes of known structure
- oxalyl-CoA
Wikimedia Foundation. 2010.