- Gla domain
Pfam_box
Symbol = Gla
Name =
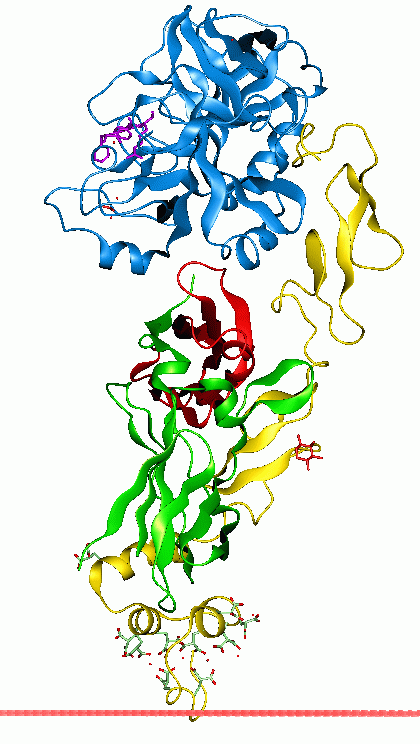
width =220
caption = Anchoring ofCoagulation factor VIIa to the membrane through its GLA domain
Pfam= PF00594
InterPro= IPR000294
SMART=
PROSITE = PDOC00011
SCOP = 1cfi
TCDB =
OPM family= 97
OPM protein= 1pfx
PDB=PDB3|1q8hA:6-47 PDB3|1q3mA:57-98 PDB3|1vzmB:53-93PDB3|1nl1A:49-90 PDB3|2spt :49-90 PDB3|1nl2A:49-90PDB3|2pf1 :79-90 PDB3|1nl8L:45-86 PDB3|1p0sL:45-86PDB3|1whf :45-86 PDB3|1whe :45-86 PDB3|1iodG:45-84PDB3|1j34C:6-46 PDB3|1j35C:6-46 PDB3|1pfxL:8-48PDB3|1cfi :52-93 PDB3|1cfh :52-93 PDB3|1mgx :52-93PDB3|1nl0G:52-91 PDB3|1lqvC:47-75 PDB3|1wv7L:65-106PDB3|1wqvL:65-106 PDB3|1wssL:65-106 PDB3|1w0yL:65-106PDB3|1z6jL:65-106 PDB3|1fakL:65-106 PDB3|1wunL:65-106PDB3|1wtgL:65-106 PDB3|1danL:65-106Vitamin K-dependent carboxylation/gamma-carboxyglutamic (GLA) domain is a
protein domain that contains post-translational modifications ofmany glutamate residues byvitamin K -dependentcarboxylation to formgamma-carboxyglutamate (Gla). The Gla residues are responsible for the high-affinity binding of calcium ions cite journal |author=Friedman PA, Przysiecki CT |title=Vitamin K-dependent carboxylation |journal=Int. J. Biochem. |volume=19 |issue=1 |pages=1–7 |year=1987 |pmid=3106112 |doi=10.1016/0020-711X(87)90116-9] cite journal |author=Vermeer C |title=Gamma-carboxyglutamate-containing proteins and the vitamin K-dependent carboxylase |journal=Biochem. J. |volume=266 |issue=3 |pages=625–636 |year=1990 |pmid=2183788] .The GLA domain is responsible for the high-affinity binding of calcium ions. It starts at the N-terminal extremity of the mature form of proteins and ends with a conserved aromatic residue; a conserved Gla-x(3)-Gla-x-Cys motifcite journal |author=Price PA, Fraser JD, Metz-Virca G |title=Molecular cloning of matrix Gla protein: implications for substrate recognition by the vitamin K-dependent gamma-carboxylase |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=84 |issue=23 |pages=8335–8339 |year=1987 |pmid=3317405 |doi=10.1073/pnas.84.23.8335] is found in the middle of the domain which seems to be important for substrate recognition by the carboxylase.
The 3D structures of several Gla domains have been solvedcite journal |author=Freedman SJ, Furie BC, Furie B, Baleja JD |title=Structure of the metal-free gamma-carboxyglutamic acid-rich membrane binding region of factor IX by two-dimensional NMR spectroscopy |journal=J. Biol. Chem. |volume=270 |issue=14 |pages=7980–7987 |year=1995 |pmid=7713897 |doi=10.1074/jbc.270.14.7980] cite journal |author=Freedman SJ, Furie BC, Furie B, Baleja JD, Blostein MD, Jacobs M |title=Identification of the phospholipid binding site in the vitamin K-dependent blood coagulation protein factor IX |journal=J. Biol. Chem. |volume=271 |issue=27 |pages=16227–16236 |year=1996 |pmid=8663165 |doi=10.1074/jbc.271.27.16227] . Calcium ions induce conformational changes in the Gla domain and are necessary for the Gla domain to fold properly. A common structural feature of functional Gla domains is the clustering of N-terminal hydrophobic residues into a hydrophobic patch that mediates interaction with the cell surface membrane.
ubfamilies
*
Coagulation factor , Gla region InterPro|IPR002383Human proteins containing this domain
BGLAP; F10; F2; F7; F9;
GAS6 ; MGP; PROC;
PROS1; PROZ;PRRG1 ;PRRG2 ;PRRG3 ;PRRG4 ;References
Wikimedia Foundation. 2010.
