- DNA laddering
-
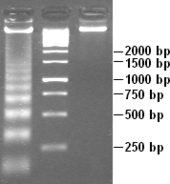 DNA laddering (left) visualised in an agarose gel by ethidium bromide staining. A 1 kb marker (middle) and control DNA (right) are included.
DNA laddering (left) visualised in an agarose gel by ethidium bromide staining. A 1 kb marker (middle) and control DNA (right) are included.
DNA laddering is a phenomenon seen in laboratory tests; it is a sensitive indicator of programmed cell death, specifically of apoptosis. It was first described in 1980 by A. H. Wyllie at the University of Edinburgh Medical School.[1]
Endonuclease activation is a characteristic feature of apoptosis. This degrades genomic DNA at internucleosomal linker regions and produces 180- to 185- base-pair DNA fragments. On Agarose gel electrophoresis, these give a characteristic "laddered" appearance. The dying cell's morphological changes are short-lived and difficult to detect. DNA laddering has therefore become a sensitive method to distinguish apoptosis from ischemic or toxic cell death.[2]
References
- ^ Wyllie AH (1980-04-10). "Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation". Nature 284 (5756): 555–556. doi:10.1038/284555a0. ISSN 0028-0836. PMID 6245367. http://www.nature.com/nature/journal/v284/n5756/abs/284555a0.html.
- ^ Iwata M, Myerson D, Torok-Storb B, Zager RA. (December 1994). "An evaluation of renal tubular DNA laddering in response to oxygen deprivation and oxidant injury". Journal of the American Society of Nephrology 5 (6): 1307–1313. ISSN 1046-6673. PMID 7893995. http://jasn.asnjournals.org/cgi/content/abstract/5/6/1307.
See also
Categories:- Apoptosis
- Biological techniques and tools
- Cell biology
- Electrophoresis
- Laboratory techniques
- Programmed cell death
- Cell biology stubs
Wikimedia Foundation. 2010.
