- Ion channel family
Pfam_box
Symbol = Ion_trans
Name = Ion channel (eukariotic)
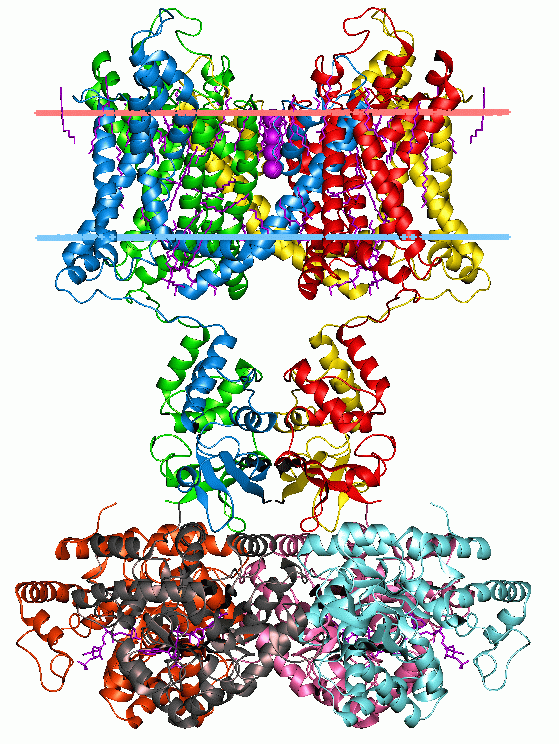
width =250
caption =Potassium channel KvAP, structure in a membrane-like environment. Calculated hydrocarbon boundaries of thelipid bilayer are indicated by red and blue dots.
Pfam= PF00520
InterPro= IPR005821
SMART=
Prosite =
SCOP = 1bl8
TCDB = 1.A.1
OPM family= 8
OPM protein= 2a79
PDB=PDB3|1qg9A:157-176 PDB3|2a79B:225-409 PDB3|1ho7A:378-397PDB3|1ho2A:378-397 PDB3|1ujlA:570-611Pfam_box
Symbol = Ion_trans_2
Name = Ion channel (bacterial)
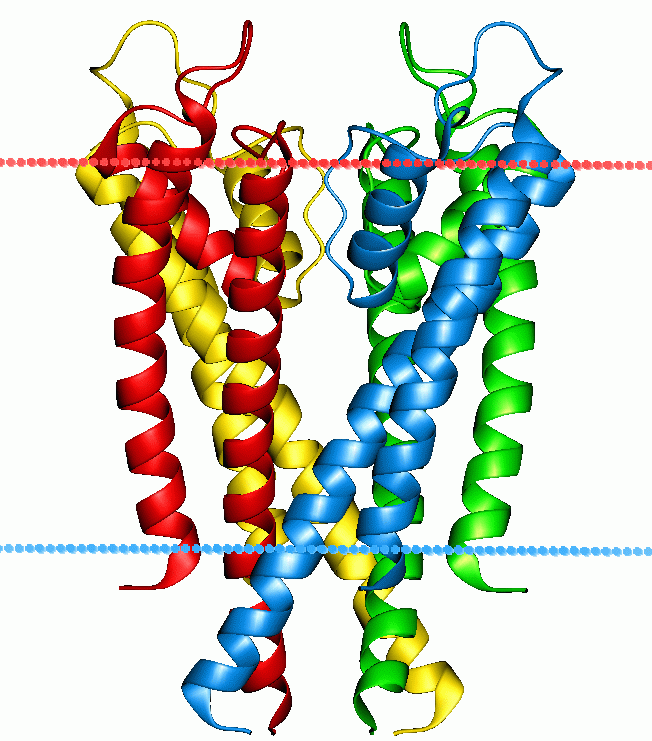
width =200
caption =Potassium channel KcsA. Calculated hydrocarbon boundaries of thelipid bilayer are indicated by red and blue dots.
Pfam= PF07885
InterPro= IPR013099
SMART=
Prosite =
SCOP = 1bl8
TCDB =
OPM family=
OPM protein= 1r3j
PDB=PDB3|1lnqE:25-100 PDB3|2a0lB:169-250 PDB3|1orqC:169-250PDB3|1k4cC:34-116 PDB3|1r3iC:34-116 PDB3|2bocC:34-116PDB3|1j95C:34-116 PDB3|1r3lC:34-116 PDB3|1jvmB:34-116PDB3|1bl8C:34-116 PDB3|2a9hD:34-116 PDB3|1k4dC:34-116PDB3|1r3jC:34-116 PDB3|1r3kC:34-116 PDB3|2bobC:34-116PDB3|1p7bB:77-151Transmembrane ion channel family was defined in
InterPro andPfam as the family of tetrameric sodium, potassium, and calciumion channels , in which two C-terminal transmembrane helices flank a loop which determinesion selectivity of the channel pore. This large group of ion channels apparently includes families TCDB|1.A.1, TCDB|1.A.2, TCDB|1.A.3, and TCDB|1.A.4 of transporter classification.Many eukariotic channels have four additional transmembrane helices (TMH) (Pfam|PF00520), whereas a bacterial structure of the protein has only two transmembrane helices that form the tetrameric channel (Pfam|PF07885).
Human channels with 6 TM helices
*
CACNA1A ;CACNA1B ;CACNA1C ;CACNA1D ;CACNA1E ;CACNA1F ;CACNA1G ;CACNA1H ;CACNA1I ;CACNA1S ;CATSPER1 ;CATSPER2 ;CATSPER3 ;CATSPER4 ;
*CNGA1 ;CNGA2 ;CNGA3 ;CNGB1 ;CNGB3 ;
*H1B ;
*HCN1 ;HCN2 ;HCN3 ;HCN4 ;HVCN1 ;
*ITPR1 ;ITPR2 ;ITPR3 ;
*KCNA1 ;KCNA10 ;KCNA2 ;KCNA3 ;KCNA4 ;KCNA5 ;KCNA6 ;KCNA7 ;KCNB1 ;KCNB2 ;KCNC1 ;KCNC2 ;KCNC3 ;KCNC4 ;KCND1 ;KCND2 ;KCND3 ;KCNF1 ;KCNG1 ;KCNG2 ;KCNG3 ;KCNG4 ;
*KCNH1 ;KCNH2 ;KCNH3 ;KCNH4 ;KCNH5 ;KCNH6 ;KCNH7 ;KCNH8 ;KCNMA1 ;
*KCNQ1 ;KCNQ2 ;KCNQ3 ;KCNQ4 ;KCNQ5 ;KCNS1 ;KCNS2 ;KCNS3 ;KCNV1 ;KCNV2 ;
*MCOLN1 ;MCOLN2 ;MCOLN3 ;MLSN1 ;NALCN ;
*PKD1L3 ;
*RCNC2 ;RYR1 ;RYR2 ;RYR3 ;
*SCN10A ;SCN11A ;SCN1A ;SCN2A ;SCN2A2 ;SCN3A ;SCN4A ;SCN5A ;SCN7A ;SCN8A ;SCN9A ;
*SLC9A10 ;SLC9A11 ;
*TPCN1 ;TPCN2 ;TRP7 ;TRPA1 ;TRPC1 ;TRPC3 ;TRPC4 ;TRPC5 ;TRPC6 ;TRPC7 ;
*TRPM1 ;TRPM2 ;TRPM3 ;TRPM4 ;TRPM5 ;TRPM6 ;TRPM7 ;TRPM8 ;
*TRPV1 ;TRPV2 ;TRPV3 ;TRPV4 ;TRPV5 ;TRPV6 ;VGCNL1 ;Human channels with two TM helices in each subunit, as in bacteria
*
KCNK1 ;KCNK10 ;KCNK12 ;KCNK13 ;KCNK15 ;KCNK16 ;KCNK17 ;KCNK18 ;KCNK2 ;KCNK3 ;KCNK4 ;KCNK5 ;KCNK6 ;KCNK7 ;KCNK9 ;KCNN1 ;KCNN2 ;KCNN3 ;KCNN4 ;
*KCNT1 ;KCNT2 ;
*SK2 ;SK3 ;SLICK ;References
* [1] . Potassium channel structures. Choe S; Nat Rev Neurosci 2002;3:115-121. PMID|11836519
Wikimedia Foundation. 2010.
