- CrcB RNA motif
-
crcB RNA motif 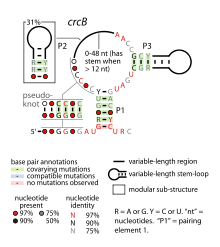
Consensus secondary structure of crcB RNAs Identifiers Symbol crcB RNA Rfam RF01734 Other data RNA type Cis-regulatory element Domain(s) Prokaryota The crcB RNA motif is a conserved RNA structure identified by bioinformatics in a wide variety of bacteria and archaea.[1] The motif exhibits the rigid or almost perfect conservation of the nucleotide identity at several positions (colored red in the diagram) across two domains of life. Such conservation is consistent with a biochemical requirement to bind a small molecule that does not evolve (like proteins or other RNAs would). Given this observation, and the fact that crcB RNAs are typically located in possible 5' untranslated regions (5' UTRs) of protein-coding genes, it was proposed that crcB RNAs likely function as riboswitches.
Many genes are presumed to be regulated by crcB RNAs. The most common are mutS, which is functions within DNA repair; the crcB gene that is thought to play a role in chromosome condensation; transporters of potassium, sodium or chloride ions; the nifU iron-sulfur protein; and genes that encode formate hydrogen lyases. Many crcB RNAs overlap predicted rho-independent transcription terminator stem-loops. Assuming that crcB RNAs are riboswitches whose conserved structure is stabilized by the interaction with their small molecule ligand, high concentrations of this ligand should destabilize the transcription terminators, and thereby lead to increased gene expression.
Only one other class of riboswitch, the TPP riboswitch, is found in organisms other than bacteria. Although several classes of RNAs are present in more than one domain of life (e.g., ribosomal RNAs, group I introns, group II introns, RNase P RNAs and HDV ribozymes), these RNAs are a small minority compared to classes of RNAs restricted to a single domain. Moreover, few of the highly widespread RNAs were initially discovered recently, while crcB RNAs were first published in 2010. The biological role played by crcB RNAs, although currently unknown, is apparently relevant to the physiology of both bacteria and archaea.
A predicted crcB RNA in Sulfolobus solfataricus P2 is contained within a non-coding transcript detected by transcriptome sequencing[2] and overlaps by 23 nucleotides an RNA called "sR110" that might correspond to a C/D box small nucleolar RNA.[3] If crcB RNAs are riboswitches, the detected transcript could reflect premature termination by riboswitch-mediated repression.
References
- ^ Weinberg Z, Wang JX, Bogue J, et al. (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571. PMID 20230605. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2864571.
- ^ Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R (January 2010). "A single-base resolution map of an archaeal transcriptome". Genome Res. 20 (1): 133–41. doi:10.1101/gr.100396.109. PMID 19884261.
- ^ Zago MA, Dennis PP, Omer AD (March 2005). "The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus". Mol. Microbiol. 55 (6): 1812–28. doi:10.1111/j.1365-2958.2005.04505.x. PMID 15752202.
Categories:- Cis-regulatory RNA elements
Wikimedia Foundation. 2010.
