- Chlorobi-1 RNA motif
-
Chlorobi-1 RNA 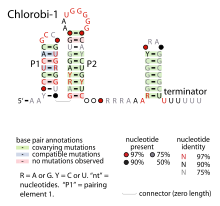
Consensus secondary structure of Chlorobi-1 RNAs Identifiers Symbol Chlorobi-1 Rfam RF01696 Other data RNA type sRNA The Chlorobi-1 RNA motif is a conserved RNA secondary structure identified by bioinformatics.[1] It is predicted to be used only by Chlorobi, a phylum of bacteria. The motif consists of two stem-loops that are followed by an apparent rho-independent transcription terminator. The motif is presumed to function as an independently transcribed non-coding RNA.
A number of other RNAs were identified in the same study, including:[1]
- Bacteroidales-1 RNA motif
- CrcB RNA Motif
- Gut-1 RNA motif
- JUMPstart RNA motif
- Lactis-plasmid RNA motif
- Lacto-usp RNA motif
- MraW RNA motif
- Ocean-V RNA motif
- PsaA RNA motif
- Pseudomon-Rho RNA motif
- Rne-II RNA motif
References
- ^ a b Weinberg Z, Wang JX, Bogue J, et al. (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571. PMID 20230605. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2864571.
External links
Categories:- Non-coding RNA
- Molecular and cellular biology stubs
Wikimedia Foundation. 2010.
