- ConsensusPathDB
-
ConsensusPathDB 
Content Description human functional interaction networks. Contact Research center Max Planck Institute for Molecular Genetics Authors Atanas Kamburov Primary Citation Kamburov & al. (2009)[1] Release date 2008 Access Data format BioPAX
PSI-MI
SBMLWebsite http://cpdb.molgen.mpg.de Tools Miscellaneous The ConsensusPathDB is a molecular functional interaction database, integrating information on protein interactions, signaling, metabolism and gene regulation in humans. ConsensusPathDB includes functional interactions from 12 databases.[1] ConsensusPathDB is freely available for academic use under http://cpdb.molgen.mpg.de/.
Contents
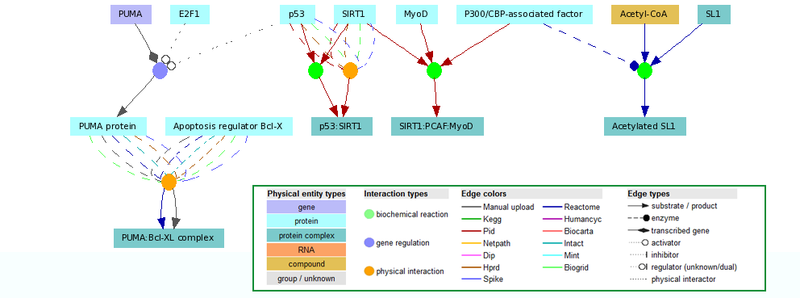
Integrated Databases
- Reactome (metabolic and signaling pathways)
- KEGG (metabolic pathways only have been integrated in ConsensusPathDB)
- HumanCyc (metabolic pathways)
- PID - Pathway Interaction Database (signaling pathways)
- BioCarta (signaling pathways)
- Netpath (signaling pathways)
- IntAct (protein interactions)
- DIP (protein interactions)
- MINT (protein interactions)
- HPRD (protein interactions)
- BioGRID (protein interactions)
- SPIKE (protein interactions, signaling reactions)
- PIG - Pathogen Interaction Gateway (host-pathogenic and host-host protein interactions)
Functionalities
The ConsensusPathDB is accessible via a web interface providing a variety of functions.
Search and visualization
Using the web interface users can search for physical entities (e.g. proteins, metabolites etc.) or pathways using common names or accession numbers (e.g. UniProt identifiers). Selected interactions can be visualized in an interactive environment as expandable networks. ConsensusPathDB currently allows users to export their models in BioPAX format or as image in several formats.
Shortest path
Users can search for shortest paths of functional interactions between physical entities, based on all interactions in the database. The pathway search can be constrained by forbidding passing through certain physical entities.
Data upload
Users can upload their own interaction networks in BioPAX, PSI-MI or SBML files in order to validate and/or extend those networks in the context of the interactions in ConsensusPathDB.
Over-representation analysis
Using the web-interface of the database, one can perform overrepresentation analysis, based on biochemical pathways or on neighbourhood-based entity sets (NESTs) that constitute sub-networks of the overall interaction network containing all physical entities around a central one within a "radius" (number of interactions from the center). For each predefined set (pathway / NEST), a P-value is computed based on the hypergeometric distribution. It reflects the significance of the observed overlap between the user-specific input gene list and the members of the predefined set.
References
- ^ a b Kamburov, Atanas; Wierling Christoph, Lehrach Hans, Herwig Ralf (Jan 2009). "ConsensusPathDB--a database for integrating human functional interaction networks" (in eng). Nucleic Acids Res. (England) 37 (Database issue): D623-8. doi:10.1093/nar/gkn698. PMC 2686562. PMID 18940869. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2686562.
External links
Categories:- Biological databases
- Systems biology
Wikimedia Foundation. 2010.