- OxyS RNA
-
OxyS RNA 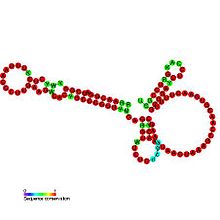
Predicted secondary structure and sequence conservation of OxyS Identifiers Symbol OxyS Rfam RF00035 Other data RNA type Gene; sRNA Domain(s) Bacteria SO 0000384 OxyS RNA is a small non-coding RNA which is induced in response to oxidative stress in Escherichia coli. This RNA acts as a global regulator to activate or repress the expression of as many as 40 genes, by an antisense mechanism, including the fhlA-encoded transcriptional activator and the rpoS-encoded sigma(s) subunit of RNA polymerase.[1] OxyS is bound by the Hfq protein, that increases the OxyS RNA interaction with its target messages.[2]. The 109 nucleotide RNA is thought to be composed of three stem-loops.
Target genes of OxyS
A number of additional targets were recently predicted and verified using microarray analysis.[3] These are listed below:
- yobF - confirmed by Northern analysis
- yaiZ
- rumA
- wrbA - confirmed by Northern analysis
- ybaY - confirmed by Northern analysis
References
- ^ Altuvia, S; Zhang A, Argaman L, Tiwari A, Storz G (1998). "The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding". EMBO J 17 (20): 6069–6075. doi:10.1093/emboj/17.20.6069. PMC 1170933. PMID 9774350. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1170933.
- ^ Zhang, A; Wassarman KM, Ortega J, Steven AC, Storz G (2002). "The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs". Mol Cell 9 (1): 11–22. doi:10.1016/S1097-2765(01)00437-3. PMID 11804582.
- ^ Tjaden B, Goodwin SS, Opdyke JA, et al. (2006). "Target prediction for small, noncoding RNAs in bacteria". Nucleic Acids Res. 34 (9): 2791–802. doi:10.1093/nar/gkl356. PMC 1464411. PMID 16717284. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1464411.
External links
Categories:- Non-coding RNA
- Molecular and cellular biology stubs
Wikimedia Foundation. 2010.
