- Oligomer restriction
-
Oligomer Restriction (abbreviated OR) is a procedure to detect an altered DNA sequence in a genome. A labeled oligonucleotide probe is hybridized to a target DNA, and then treated with a restriction enzyme. If the probe exactly matches the target, the restriction enzyme will cleave the probe, changing its size. If, however, the target DNA does not exactly match the probe, the restriction enzyme will have no effect on the length of the probe. The OR technique, now rarely performed, was closely associated with the development of the popular polymerase chain reaction (PCR) method.
Contents
Example
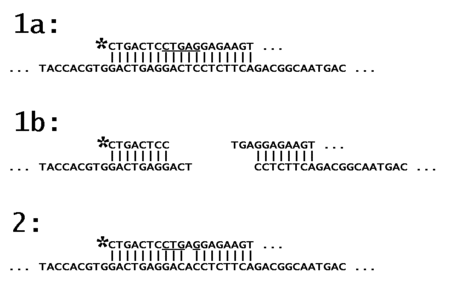
In part 1a of the schematic the oligonucleotide probe, labeled on its left end (asterisk), is shown on the top line. It is fully complementary to its target DNA (here taken from the human β-hemoglobin gene), as shown on the next line. Part of the probe includes the Recognition site for the restriction enzyme Dde I (underlined).
In part 1b, the restriction enzyme has cleaved the probe and its target (Dde I leaves three bases unpaired at each end). The labeled end of the probe is now just 8 bases in length, and is easily separated by Gel electrophoresis from the uncut probe, which was 40 bases long.
In part 2, the same probe is shown hybridized to a target DNA which includes a single base mutation (here the mutation responsible for Sickle Cell Anemia, or SCA). The mismatched hybrid no longer acts as a recognition site for the restriction enzyme, and the probe remains at its original length.
History
The Oligomer Restriction technique was developed as a variation of the Restriction Fragment Length Polymorphism (RFLP) assay method, with the hope of avoiding the laborious Southern blotting step used in RFLP analysis. OR was conceived by Randall Saiki and Henry Erlich in the early 1980s, working at Cetus Corporation in Emeryville, California. It was patented in 1984[1] and published in 1985[2], having been applied to the genomic mutation responsible for Sickle Cell Anemia. OR was soon replaced by the more general technique of Allele Specific Oligonucleotide (ASO) probes[3].
Problems
The Oligomer Restriction method was beset by a number of problems:
- It could be applied only to the small set of DNA polymorphisms which alter a restriction site, and only to those sites for which sequence information was known. Many of the known RFLP assays detected polymorphisms which were far away from their probe locations.
- It is difficult to label oligonucleotides to a level high enough to use them as probes for genomic DNA. This problem also plagued the development of ASO probes.
- It is difficult to design oligonucleotides and use them in a way that they become hybridization probes for just a single site within a genome. Binding to non-specific locations can often obscure the effect of the probe at the target location.
- Not all restriction enzymes have the desired specificity for their recognition sequence. Some can recognize and cut single-stranded DNA, and some show a low level of cleavage for mismatched sites. Even a small amount of non-specific cleavage can swamp the weak signal expected from the target sequence.
- It was difficult to design an OR method that included controls for both of the alleles being tested. In part 2 of the simplified example described above, the probe was not cleaved when hybridized to a mutant target. But the same (non-) result would occur for the large excess of unhybridized probe, as well as if any problem occurred preventing the complete digestion by restriction enzyme. In the actual method reported[2], a second non-polymorphic restriction site was used to cut all of the hybridized probe, and a second unlabeled oligonucletide was used to 'block' the unhybridized probe. These controls would not have been available for other targets.
Relationship to PCR
Despite its limitations, the OR technique benefited from its close association with the development of the polymerase chain reaction. Kary Mullis, who also worked at Cetus, had synthesized the oligonucleotide probes being tested by Saiki and Erlich. Aware of the problems they were encountering, he envisioned an alternative method for analyzing the SCA mutation that would use components of the Sanger DNA sequencing technique. Realizing the difficulty of hybridizing an oligonucleotide primer to a single location in the genome, he considered using a second primer on the opposite strand. He then generalized that process and realized that repeated extensions of the two primers would lead to an exponential increase in the segment of DNA between the primers - a chain reaction of replication catalyzed by DNA polymerase[4][5].
As Mullis encountered his own difficulties in demonstrating PCR[6], he joined an existing group of researchers that were addressing the problems with OR. Together, they developed the combined PCR-OR assay. Thus, OR became the first method used to analyze PCR-amplified genomic DNA.
Mullis also encountered difficulties in publishing the basic idea of PCR (scientific journals rarely publish concepts without accompanying results). When his manuscript for the journal Nature was rejected, the basic description of PCR was hurriedly added to the paper originally intended to report the OR method (Mullis was also a co-author there). This OR paper[2] thus became the first publication of PCR, and for several years would become the report most cited by other researchers.
References
- ^ Saiki RK, Erlich, HA "Method for detection of polymorphic restriction sites and nucleic acid sequences." U.S. Patent 4683194.
- ^ a b c Saiki, RK; Scharf S, Faloona F, Mullis KB, Erlich HA, Arnheim N (December 20 1985). "Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia". Science 230 (4732): 1350–4. doi:10.1126/science.2999980. PMID 2999980. http://sunsite.berkeley.edu/cgi-bin/ebind2html/pcr/034.
- ^ Saiki RK, Bugawan TL, Mullis KB, and Erlich HE "Analysis of enzymatically amplified beta-globin and HLA-DQa DNA with allele-specific oligonucleotide probes" Nature vol. 324(6093) pp. 163-166 (1986).
- ^ Mullis K "The Unusual Origin of the Polymerase Chain Reaction" Scientific American vol. 262(4): pp. 56-65 (1990).
- ^ Mullis K "The Polymerase Chain Reaction", Nobel Lecture, December 8, 1993.
- ^ Rabinow P "Making PCR: A Story of Biotechnology" University of Chicago Press (1996).
Categories:- DNA profiling techniques
- Molecular biology techniques
- Molecular biology
- Laboratory techniques
Wikimedia Foundation. 2010.