- Sema domain
Pfam_box
Symbol = Sema
Name =
width =250
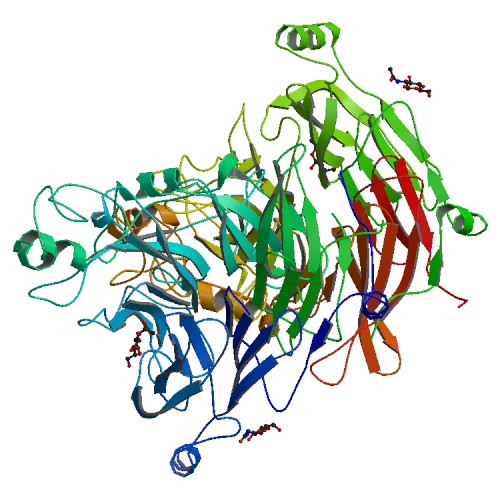
caption =Sema domain, immunoglobulin domain (Ig), short basic domain
Pfam= PF01403
InterPro= IPR001627
SMART=
PROSITE = PDOC51004
SCOP = 1olz
TCDB =
OPM family=
OPM protein=
PDB=PDB3|1shyB:55-500 PDB3|1ux3A:55-500 PDB3|1olzA:50-482PDB3|1q47A:57-498The Sema domain is a
structural domain ofsemaphorin s, which are a large family of secreted and transmembrane proteins, some of which function as repellent signals duringaxon guidance . Sema domains also occur in UniProt|P08581 the hepatocyte growth factor receptor and UniProt|P51805 cite journal |author=Goodman CS, Winberg ML, Noordermeer JN, Tamagnone L, Comoglio PM, Spriggs MK, Tessier-Lavigne M |title=Plexin A is a neuronal semaphorin receptor that controls axon guidance |journal=Cell |volume=95 |issue=7 |pages=903–916 |year=1998 |pmid=9875845 |doi=10.1016/S0092-8674(00)81715-8] and in viral proteins.CD100 (also called SEMA4D) is associated with
PTPase andserine kinase activity. CD100 increases PMA, CD3 and CD2 induced T cell proliferation, increases CD45 induced T cell adhesion, induces B cell homotypic adhesion and down-regulatesB cell expression ofCD23 .The Sema domain is characterised by a conserved set of cysteine residues, which form four disulfide bonds to stabilise the structure. The Sema domain fold is a variation of the beta propeller topology, with seven blades radiallyarranged around a central axis. Each blade contains a four- stranded (strands A to D) antiparallel beta sheet. The inner strand of each blade (A) lines the channel at the centre of the propeller, with strands B and C of the same repeat radiating outward, and strand D of the next repeat forming the outer edge of the blade. The large size of the Sema domain is not due to a single inserted domain but results from the presence of additional secondary structure elements inserted in most of the blades. The Sema domain uses a 'loop and hook' system to close the circle between the first and the last blades. The blades are constructed sequentially with an N-terminal beta- strand closing the circle by providing the outermost strand (D) of the seventh (C-terminal) blade. The beta-propeller is further stabilized by an extension of the N-terminus, providing an additional, fifth beta-strand on the outer edge of blade 6cite journal |author=Nikolov DB, Himanen JP, Rajashankar KR, Lu M, Antipenko A, Lesniak J, Barton WA, Hoemme C, Puschel AW, van Leyen K, Nardi-Dei V |title=Structure of the semaphorin-3A receptor binding module |journal=Neuron |volume=39 |issue=4 |pages=589–598 |year=2003 |pmid=12925274 |doi=10.1016/S0896-6273(03)00502-6] cite journal |author=Stuart DI, Jones EY, Harlos K, Esnouf RM, Davis SJ, Love CA, Mavaddat N |title=The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D |journal=Nat. Struct. Biol. |volume=10 |issue=10 |pages=843–848 |year=2003 |pmid=12958590 |doi=10.1038/nsb977] cite journal |author=Lazarus RA, Kirchhofer D, Stamos J, Yao X, Wiesmann C |title=Crystal structure of the HGF beta-chain in complexwith the Sema domain of the Met receptor |journal=EMBO J. |volume=23 |issue=12 |pages=2325–2335 |year=2004 |pmid=15167892 |doi=10.1038/sj.emboj.7600243] .
CD molecules are leucocyte antigens on cell surfaces. CD antigens nomenclature is updated at Protein Reviews On The Web (http://mpr.nci.nih.gov/prow/).
Human proteins containing this domain
MET ;MST1R ;PLXNA1 ;PLXNA2 ;PLXNA3 ;PLXNA4 ;PLXNB1 ;PLXNB2 ;PLXNB3 ;PLXND1 ;SEMA3A ;SEMA3B ;SEMA3C ;SEMA3D ;SEMA3E ;SEMA3F ;SEMA3G ;SEMA4A ;SEMA4B ;SEMA4C ;SEMA4D ;SEMA4F ;SEMA4G ;SEMA5A ;SEMA5B ;SEMA6A ;SEMA6B ;SEMA6C ;SEMA6D ;SEMA7A ;References
Wikimedia Foundation. 2010.
