- Dam (methylase)
-
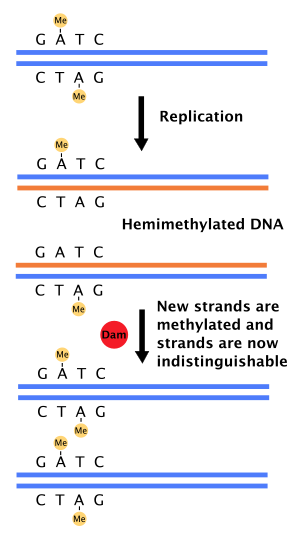
DAM methylase (DNA adenine methylase) is an enzyme that adds a methyl group to the adenine of the sequence 5'-GATC-3' in newly synthesized DNA.[1][2] Immediately after DNA synthesis, the daughter strand remains unmethylated for a short time.[3]
Contents
Role in mismatch repair of DNA
When DNA polymerase makes an error resulting in a mismatched base-pair or a small insertion or deletion during DNA synthesis, the cell will repair the DNA by a pathway called mismatch repair. However, the cell must be able to differentiate between the template strand and the newly synthesized strand. In bacteria, DNA strands are methylated by Dam methylase, and therefore, immediately after replication, the DNA will be hemimethylated.[3] A repair enzyme, MutS, binds to mismatches in DNA and recruits MutL, which subsequently activates the endonuclease MutH. MutH binds hemimethylated GATC sites and when activated will selectively cleave the unmethylated daughter strand, allowing helicase and exonucleases to excise the nascent strand in the region surrounding the mismatch.[3][4] The strand is then re-synthesized by DNA polymerase III.
Role in regulation of replication
The firing of the origin of replication (oriC) in bacteria cells is highly controlled to ensure DNA replication occurs only once during each cell division. Part of this can be explained by the slow hydrolysis of ATP by DnaA, a protein that binds to repeats in the oriC to initiate replication. Dam methylase also plays a role because the oriC has 11 5'-GATC-3' sequences (in E. coli). Immediately after DNA replication, the oriC is hemimethylated and sequestered for a period of time. Only after this, the oriC is released and must be fully methylated by Dam methylase before DnaA binding occurs.
Role in regulation of protein expression
Dam also plays a role in the promotion and repression of RNA transcription. In E. coli downstream GATC sequences are methylated, promoting transcription. For example, Pap pilus phase variation in uropathogenic E. coli is controlled by Dam through the methylation of the two GATC sites proximal and distal to the pap promoter.[5]
See also
- Dam dcm strain
- DNA replication
- methylase
- mismatch repair
References
- ^ Marinus MG, Morris NR (June 1973). "Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12". J. Bacteriol. 114 (3): 1143–50. PMC 285375. PMID 4576399. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=285375.
- ^ Geier GE, Modrich P (February 1979). "Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease". J. Biol. Chem. 254 (4): 1408–13. PMID 368070.
- ^ a b c Barras F, Marinus MG (1989). "The great GATC: DNA methylation in E. coli". Trends in Genetics 5: 139–143. doi:10.1016/0168-9525(89)90054-1.
- ^ Løbner-Olesen A, Skovgaard O, Marinus, MG (April 2005). "Dam methylation: coordinating cellular processes". Curr. Opin. Microbiol. 8 (2): 154–160. doi:10.1016/j.mib.2005.02.009. PMID 15802246.
- ^ Casadesús J, Low D (September 2006). "The great GATC: DNA methylation in E. coli". Microbiology and Molecular Biology Reviews 70 (3): 830–856. doi:10.1128/MMBR.00016-06. PMC 1594586. PMID 16959970. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1594586.
External links
Categories:- EC 2.1.1
Wikimedia Foundation. 2010.