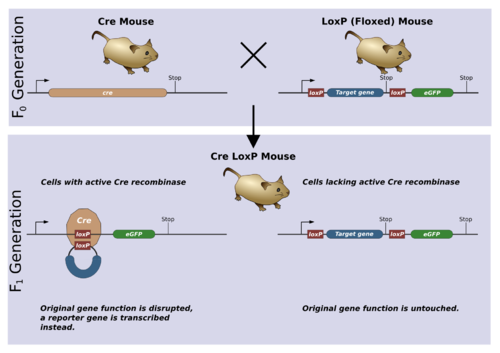
- Cre-Lox recombination
-
Cre-Lox recombination is a special type of site-specific recombination developed by Dr. Brian Sauer initially for use in activating gene expression in mammalian cell lines and transgenic mice (DuPont).[1][2] Subsequently, the laboratory of Dr. Jamey Marth showed that Cre-Lox recombination could be used to delete loxP-flanked chromosomal DNA sequences at high efficiency in selected cell types of transgenic animals, suggesting this approach as a means to define gene function in specific cell types, indelibly mark progenitors in cell fate determination studies, induce specific chromosomal rearrangements for biological and disease modeling, and determine the roles of early genetic lesions in disease (and phenotype) maintenance.[3]. Shortly thereafter, the laboratory of Dr. Klaus Rajewsky reported the production of pluripotent embryonic stem cells bearing a targeted loxP-flanked (floxed) DNA polymerase gene.[4] Combining these advances in collaboration, the laboratories of Drs. Marth and Rajewsky showed in 1994 that Cre-lox recombination could be used for conditional gene targeting in vivo. [5] This technique continues to be used by hundreds of researchers and laboratories around the world as an essential procedure to discover gene function in normal and disease biology, resulting in numerous important discoveries that would otherwise have not been possible. In 2009, Germany awarded the Max Delbrück medal to Dr. Klaus Rajewsky for his role in developing Cre-Lox recombination. Cre-Lox recombination involves the targeting of a specific sequence of DNA and splicing it with the help of an enzyme called Cre recombinase. Because systemic inactivation of many genes further cause embryonic lethality, Cre-Lox recombination is commonly used to circumvent this problem. In addition, Cre–Lox recombination provides the best experimental control that presently exists in transgenic animal modeling to link genotypes (alterations in genomic DNA) to the biological outcomes (phenotypes).
Contents
Overview
The Cre-lox system is used as a genetic tool to control site specific recombination events in genomic DNA. This system has allowed researchers to manipulate a variety of genetically modified organisms to control gene expression, delete undesired DNA sequences and modify chromosome architecture.
The Cre protein is a site-specific DNA recombinase, that is, it can catalyse the recombination of DNA between specific sites in a DNA molecule. These sites, known as loxP sequences, contain specific binding sites for Cre that surround a directional core sequence where recombination can occur.
When cells that have loxP sites in their genome express Cre, a recombination event can occur between the loxP sites. The double stranded DNA is cut at both loxP sites by the Cre protein. The strands are then rejoined with DNA ligase in a quick and efficient process. The result of recombination depends on the orientation of the loxP sites. For two lox sites on the same chromosome arm, inverted loxP sites will cause an inversion of the intervening DNA, while a direct repeat of loxP sites will cause a deletion event. If loxP sites are on different chromosomes it is possible for translocation events to be catalysed by Cre induced recombination.
Cre recombinase
The Cre (causes recombination)[6] protein consists of 4 subunits and two domains: The larger carboxyl (C-terminal) domain, and smaller amino (N-terminal) domain. The total protein has 343 amino acids. The C domain is similar in structure to the domain in the Integrase family of enzymes isolated from lambda phage. This is also the catalytic site of the enzyme.
Lox P site
Lox P (locus of X-over P1) is a site on the Bacteriophage P1 consisting of 34 bp. There exists an asymmetric 8 bp sequence in between with two sets of palindromic, 13 bp sequences flanking it. The detailed structure is given below.
13bp 8bp 13bp ATAACTTCGTATA - GCATACAT -TATACGAAGTTAT Holliday junctions and homologous recombination
During genetic recombination, a Holliday junction is formed between the two strands of DNA and a double-stranded break in a DNA molecule leaves a 3’OH end exposed. This reaction is aided with the endonuclease activity of an enzyme. 5’ Phosphate ends are usually the substrates for this reaction, thus extended 3’ regions remain. This 3’ OH group is highly unstable, and the strand on which it is present must find its complement. Since Homologous Recombination occurs after DNA replication, two strands of DNA are available, and, thus, the 3’ OH group must pair with its complement, and it does so, with an intact strand on the other duplex. Now, one point of crossover has occurred, which is what is called a Holliday Intermediate.
The 3’OH end is elongated (that is, bases are added) with the help of DNA Polymerase. The pairing of opposite strands is what constitutes the crossing-over or Recombination event, which is common to all living organisms, since the genetic material on one strand of one duplex has paired with one strand of another duplex, and has been elongated by DNA polymerase. Further cleavage of Holliday Intermediates results in formation of Hybrid DNA.
This further cleavage or ‘resolvation’ is done by a special group of enzymes called Resolvases. RuvC is just one of these Resolvases that have been isolated in bacteria and yeast.
For many years, it was thought that, when the Holliday junction intermediate was formed, the branch point of the junction (where the strands cross over) would be located at the first cleavage site. Migration of the branch point to the second cleavage site would then somehow trigger the second half of the pathway. This model provided convenient explanation for the strict requirement for homology between recombining sites, since branch migration would stall at a mismatch and would not allow the second strand exchange to occur. In more recent years, however, this view has been challenged, and most of the current models for Int, Xer, and Flp recombination involve only limited branch migration 1–3 base pairs) of the Holliday intermediate, coupled to an isomerisation event that is responsible for switching the strand cleavage specificity.
Site-specific recombination
Site-specific recombination (SSR) involves specific sites for the catalysing action of special enzymes called recombinases. Cre, or cyclic recombinase, is one such enzyme. Site-specific recombination is, thus, the enzyme-mediated cleavage and ligation of two defined deoxynucleotide sequences.
A number of conservative site-specific recombination systems have been described in both prokaryotic and eukaryotic organisms. In general, these systems use one or more proteins and act on unique asymmetric DNA sequences. The products of the recombination event depend on the relative orientation of these asymmetric sequences. Many other proteins apart from the Recombinase are involved in regulating the reaction. During site-specific DNA recombination, which brings about genetic rearrangement in processes such as viral integration and excision and chromosomal segregation, these recombinase enzymes recognize specific DNA sequences and catalyse the reciprocal exchange of DNA strands between these sites.
Mechanism of action
Initiation of site-specific recombination begins with the binding of recombination proteins to their respective DNA targets. A separate recombinase recognizes and binds to each of two recombination sites on two different DNA molecules or within the same DNA. At the given specific site on the DNA, the hydroxyl group of the tyrosine attacks a phosphate group in the DNA using a direct transesterification mechanism linking the recombinase protein to the DNA via a phospho-tyrosine linkage. This conserves the energy of the phosphodiester bond, allowing the reaction to be reversed without the involvement of a high-energy cofactor.
Cleavage on the other strand also causes a phospho-tyrosine bond between DNA and the enzyme. At both the DNA duplexes, the bonding of the phosphate group to tyrosine residues leave a 3’ OH group free. In fact, the enzyme-DNA complex is an intermediate stage, which is followed by the ligation of the 3’ OH group of one DNA strand to the 5’ phosphate group of the other DNA strand, which is covalently bonded to the tyrosine residue; that is, the covalent linkage between 5’ end and tyrosine residue is broken. This reaction synthesizes the Holliday Junction discussed earlier.
In this fashion, opposite DNA strands are joined together. Subsequent cleavage and rejoining cause DNA strands to exchange their segments. Protein-Protein interactions drive and direct strand exchange. Energy is not compromised, since the Protein-DNA linkage makes up for the loss of the Phosphodiester bond, which occurred during cleavage.
Site-specific Recombination is also an important process that viruses, such as bacteriophages, adopt to integrate their genetic material into the infected host. The virus, called a prophage in such a state, accomplishes this via integration and excision. The points where the integration and excision reactions occur are called the attachment (att) sites. An attP site on the Phage exchanges segments with an attB site on the Bacterial DNA. Thus, these are site-specific, occurring only at the respective att sites. The integrase class of enzymes catalyse this particular reaction.
The Cre-lox system in action
In initial studies in the Bacteriophage P1 system, it was noticed that the gene to be excised was flanked (sandwiched) by two loxP DNA sequences or sites. The presence of the Cre enzyme resulted in the Phage P1 chromosome's assuming a structure that allowed the two loxP sites to come in such close contact that the site-specific recombination mechanism (described above) was capable of taking place. This site-specific recombination resulted in the excision of the flanked gene or DNA sequence from the P1 chromosome into a circular structure.
Implementation of multiple loxP site pairs
Multiple variants of loxP,[7] in particular lox2272 and loxN, have been used by researchers with the combination of different Cre actions (transient or constitutive) to create a "Brainbow" system that allows multi-colouring of mice's brain with four fluorescent proteins.
References
- ^ Sauer, B. (1987) "Functional expression of the Cre-Lox site-specific recombination system in the yeast Saccharomyces cerevisiae", Mol Cell Biol 7: 2087-2096
- ^ Sauer, B. and Henderson, N. (1988) "Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1." Proc. Natl. Acad. Sci. USA, 85: 5166-5170
- ^ Orban, P.C., Chui, D., and Marth, J.D. (1992) "Tissue– and site–specific recombination in transgenic mice." "Proc. Natl. Acad. Sci. USA" "89": 6861–6865
- ^ Gu, H., Zou, Y.R., and Rajewsky, K. (1993) "Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre–loxP–mediated gene targeting." "Cell" "73": 1155-1564
- ^ Gu, H., Marth, J.D., Orban, P.C., Mossman, H., and Rajewsky, K. (1994) "Deletion of the DNA polymerase beta gene in T cells using tissue-specific gene targeting. "Science" "265": 103-106.
- ^ Sternberg and Hamilton, J.Mol.Biol. (1981) 150, 467-486, pp 468
- ^ Editor's Summary (1 November 2007) Over the brainbow. Nature. Accessed 15 March 2010.
External links
Categories:
Wikimedia Foundation. 2010.