- Force field (chemistry)
In the context of
molecular mechanics , a force field (also called a forcefield) refers to the functional form andparameter sets used to describe thepotential energy of a system of particles (typically but not necessarilyatom s). Force field functions and parameter sets are derived from both experimental work and high-level quantum mechanical calculations. "All-atom" force fields provide parameters for every atom in a system, includinghydrogen , while "united-atom" force fields treat the hydrogen andcarbon atoms inmethyl andmethylene groups as a single interaction center. "Coarse-grained" force fields, which are frequently used in long-time simulations ofprotein s, provide even more abstracted representations for increased computational efficiency.The usage of the term "force field" in chemistry and computational biology differs from the standard usage in
physics . In chemistry usage a force field is defined as a potential function, while the term is used in physics to denote the negativegradient of ascalar potential .Functional form
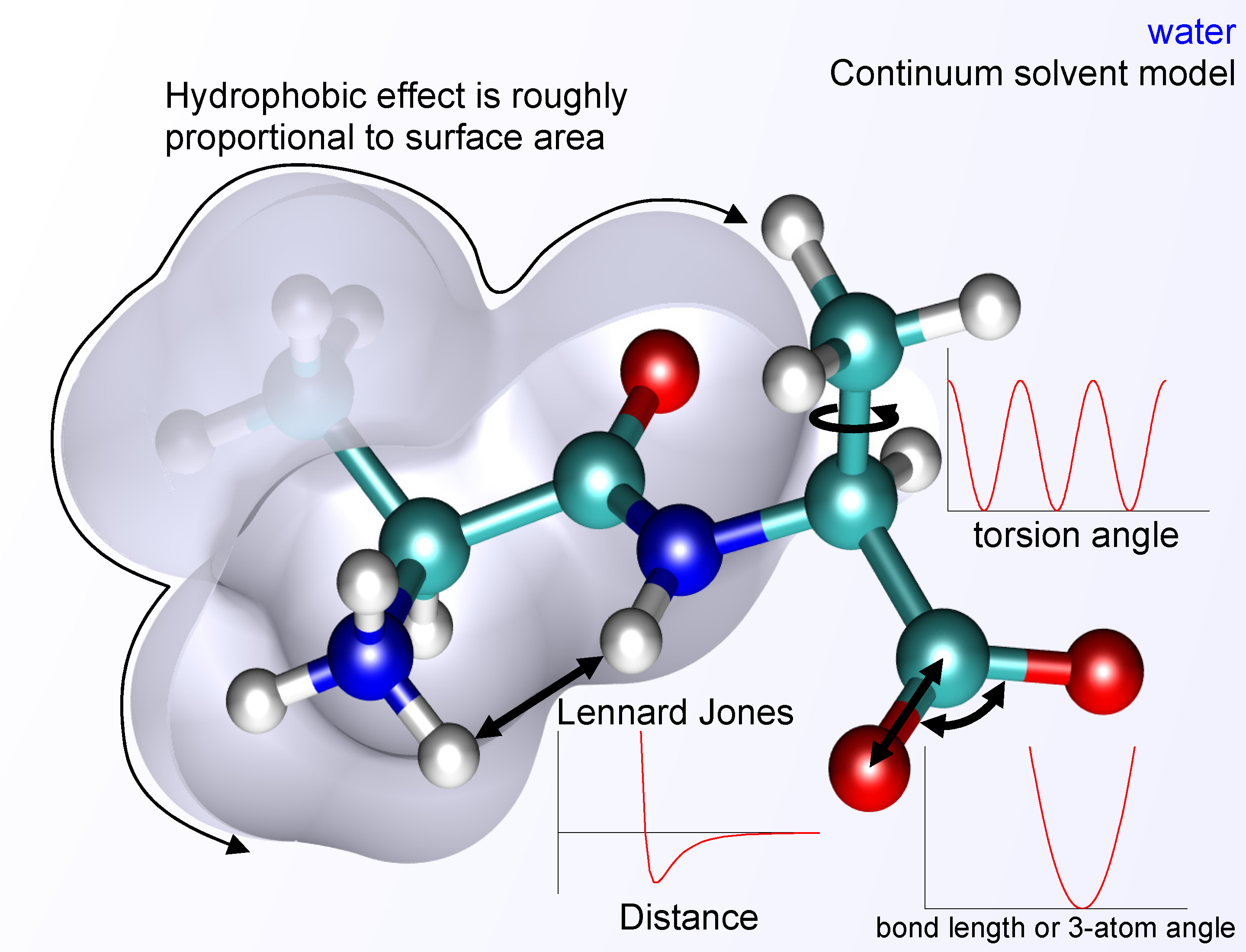
thumb|right|Molecular mechanics potential energy function with continuum solvent.The basic functional form of a force field encapsulates both bonded terms relating to atoms that are linked bycovalent bond s, and nonbonded (also called "noncovalent") terms describing the long-rangeelectrostatic and van der Waals forces. The specific decomposition of the terms depends on the force field, but a general form for the total energy in an additive force field can be written as where the components of the covalent and noncovalent contributions are given by the following summations:The bond and angle terms are usually modeled as
harmonic oscillator s in force fields that do not allow bond breaking. A more realistic description of a covalent bond at higher stretching is provided by the more expensiveMorse potential . The functional form for the rest of the bonded terms is highly variable. Proper dihedral potentials are usually included. Additionally, "improper torsional" terms may be added to enforce the planarity ofaromatic rings and otherconjugated system s, and "cross-terms" that describe coupling of different internal variables, such as angles and bond lengths. Some force fields also include explicit terms forhydrogen bond s.The nonbonded terms are most computationally intensive because they include many more interactions per atom. A popular choice is to limit interactions to pairwise energies. The van der Waals term is usually computed with a
Lennard-Jones potential and the electrostatic term withCoulomb's law , although both can be buffered or scaled by a constant factor to account for electronicpolarizability and produce better agreement with experimental observations.Parameterization
In addition to the functional form of the potentials, a force field defines a set of parameters for each type of atom. For example, a force field would include distinct parameters for an
oxygen atom in acarbonyl functional group and in ahydroxyl group. The typical parameter set includes values foratomic mass ,van der Waals radius , andpartial charge for individual atoms, and equilibrium values ofbond length s, bond angles, anddihedral angle s for pairs, triplets, and quandruplets of bonded atoms, and values corresponding to the effectivespring constant for each potential. Most current force fields use a "fixed-charge" model by which each atom is assigned a single value for the atomic charge that is not affected by the localelectrostatic environment; proposed developments in next-generation force fields incorporate models forpolarizability , in which a particle's charge is influenced by electrostatic interactions with its neighbors. For example, polarizability can be approximated by the introduction of induced dipoles; it can also be represented byDrude particle s, or massless, charge-carrying virtual sites attached by a springlike harmonic potential to each polarizable atom. The introduction of polarizability into force fields in common use has been inhibited by the high computational expense associated with calculating the local electrostatic field.Although many molecular simulations involve biological
macromolecule s such asprotein s,DNA , andRNA , the parameters for given atom types are generally derived from observations on small organic molecules that are more tractable for experimental studies and quantum calculations. Different force fields can be derived from dissimilar types of experimental data, such asenthalpy ofvaporization (OPLS ),enthalpy ofsublimation (CFF ),dipole moment s, or various spectroscopic parameters (CFF).Parameter sets and functional forms are defined by force field developers to be
self-consistent . Because the functional forms of the potential terms vary extensively between even closely related force fields (or successive versions of the same force field), the parameters from one force field should never be used in conjunction with the potential from another.Deficiencies
All force fields are based on numerous approximations and derived from different types of experimental data. Therefore they are called "empirical". Some existing force fields usually do not account for electronic
polarization of the environment, an effect that can significantly reduce electrostatic interactions of partial atomic charges. This problem was addressed by developing “polarizable force fields” Ponder JW and Case DA. (2003) Force fields for protein simulations. "Adv. Prot. Chem." 66: 27-85.] Warshel A, Sharma PK, Kato M and Parson WW (2006) Modeling Electrostatic Effects in Proteins. Biochim. Biophys. Acta 1764:1647-1676.] or using macroscopicdielectric constant . However, application of a single value ofdielectric constant is questionable in the highly heterogeneous environments of proteins or biological membranes, and the nature of the dielectric depends on the model used Schutz CN. and Warshel A. 2001. What are the dielectric "constants" of proteins and how to validate electrostatic models? Proteins 44: 400-417. ] .All types of
van der Waals force s are also strongly environment-dependent, because these forces originate from interactions of induced and “instantaneous” dipoles (seeIntermolecular force ). The originalFritz London theory of these forces can only be applied in vacuum. A more general theory ofvan der Waals force s in condensed media was developed by A. D. McLachlan in 1963 (this theory includes the original London’s approach as a special case) Israelachvili, J.N. 1992. "Intermolecular and surface forces." Academic Press, San Diego.] . The McLachlan theory predicts that van der Waals attractions in media are weaker than in vacuum and follow the "like dissolves like" rule, which means that different types of atoms interact more weakly than identical types of atoms. Leckband, D. and Israelachvili, J. (2001) Intermolecular forces in biology. "Quart. Rev. Biophys." 34: 105-267.] . This is in contrast to “combinatorial rules” or Slater-Kirkwood equation applied for development of the classical force fields. The “combinatorial rules” state that interaction energy of two dissimilar atoms (e.g. C…N) is an average of the interaction energies of corresponding identical atom pairs (i.e. C…C and N…N). According to McLachlan theory, the interactions of particles in a media can even be completely repulsive, as observed for liquidhelium . The conclusions of McLachlan theory are supported by direct measurements of attraction forces between different materials (Hamaker constant), as explained byJacob Israelachvili in his book "Intermolecular and surface forces". It was concluded that "the interaction between hydrocarbons across water is about 10% of that across vacuum" . Such effects are unaccounted in the standard molecular mechanics.Another round of criticism came from practical applications, such as protein structure refinement. It was noted that
CASP participants did not try to refine their models to avoid "a central embarrassment of molecular mechanics, namely that energy minimization or molecular dynamics generally leads to a model that is less like the experimental structure". Koehl P. and Levitt M. (1999) A brighter future for protein structure prediction. "Nature Struct. Biol." 6: 108-111.] Actually, the force fields have been successfully applied for protein structure refinement in differentX-ray crystallography andNMR spectroscopy applications, especially using program XPLOR Brunger AT and Adams PD. (2002) Molecular dynamics applied to X-ray structure refinement. "Acc. Chem. Res." 35: 404-412.] . However, such refinement is driven primarily by a set of experimental constraints, whereas the force fields serve merely to remove interatomic hindrances. The results of calculations are practically the same with rigid sphere potentials implemented in program DYANA Guntert P. (1998) Structure calculation of biological macromolecules from NMR data. "Quart. Rev. Biophys." 31: 145-237.] (calculations from NMR data), or with programs for crystallographic refinement that do not use any energy functions. The deficiencies of the force fields remain a major bottleneck inhomology modeling of proteins Tramontano A. and Morea V. 2003. Assessment of homology-based predictions in CASP5. "Proteins." 53: 352-368.] . Such situation gave rise to development of alternative empirical scoring functions specifically forligand docking Gohlke H. and Klebe G. (2002) Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. "Angew. Chem. Internat. Ed." 41: 2644-2676. ] ,protein folding Edgcomb SP. and Murphy KP. (2000) Structural energetics of protein folding and binding. "Current Op. Biotechnol." 11: 62-66.] Lazaridis T. and Karplus (2000) Effective energy functions for protein structure prediction. "Curr. Op. Struct. Biol." 10: 139-145] Levitt M. and Warshel A. (1975) Computer Simulations of Protein Folding, Nature 253: 694-698] , computationalprotein design Gordon DB, Marshall SA, and Mayo SL (1999) Energy functions for protein design. "Curr. Op. Struct. Biol." 9: 509-513.] Mendes J., Guerois R, and Serrano L (2002) Energy estimation in protein design. "Curr. Op. Struct. Biol." 12: 441-446. ] Rohl CA, Strauss CEM, Misura KMS, and Baker D. (2004) Protein structure prediction using Rosetta. "Meth. Enz." 383: 66-93. ] , and modeling of proteins in membranes Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI (2006) Positioning of proteins in membranes: A computational approach. "Protein Sci." 15, 1318-1333.] .There is also an opinion that molecular mechanics may operate with energy which is irrelevant to protein folding or ligand binding Lomize A.L., Reibarkh M.Y. and Pogozheva I.D. (2002) Interatomic potentials and solvation parameters from protein engineering data for buried residues. "Protein Sci.", 11:1984-2000.] . The parameters of typical force fields reproduce
enthalpy of sublimation, i.e. energy of evaporation of molecular crystals. However, it was recognized thatprotein folding andligand binding are thermodynamically very similar tocrystallization , or liquid-solid transitions, because all these processes represent “freezing” of mobile molecules in condensed media Murphy K.P. and Gill S.J. 1991. Solid model compounds and the thermodynamics of protein unfolding. "J. Mol. Biol.", 222: 699-709.] Shakhnovich, E.I. and Finkelstein, A.V. (1989) Theory of cooperative transitions in protein molecules. I. Why denaturation of globular proteins is a first-order phase transition. "Biopolymers" 28: 1667-1680. ] Graziano, G., Catanzano, F., Del Vecchio, P., Giancola, C., and Barone, G. (1996) Thermodynamic stability of globular proteins: a reliable model from small molecule studies. "Gazetta Chim. Italiana" 126: 559-567.] . Therefore, free energy changes during protein folding or ligand binding are expected to represent a combination of an energy similar toheat of fusion (energy absorbed during melting of molecular crystals), aconformational entropy contribution, andsolvation free energy. Theheat of fusion is significantly smaller than enthalpy of sublimation . Hence, the potentials describing protein folding or ligand binding must be weaker than potentials in molecular mechanics. Indeed, the energies ofH-bond s in proteins are ~ -1.5 kcal/mol when estimated fromprotein engineering oralpha helix to coil transition data Myers J.K. and Pace C.N. (1996) Hydrogen bonding stabilizes globular proteins, "Biophys. J." 71: 2033-2039. ] Scholtz J.M., Marqusee S., Baldwin R.L., York E.J., Stewart J.M., Santoro M., and Bolen D.W. (1991) Calorimetric determination of the enthalpy change for the alpha-helix to coil transition of an alanine peptide in water. "Proc. Natl. Acad. Sci. USA" 88: 2854-2858. ] , but the same energies estimated from sublimationenthalpy of molecularcrystal s were -4 to -6 kcal/mol Gavezotti A. and Filippini G. (1994) Geometry of intermolecular X-H...Y (X,Y=N,O) hydrogen bond and the calibration of empirical hydrogen-bond potentials. "J. Phys. Chem." 98: 4831-4837.] . The depths of modifiedLennard-Jones potential s derived from protein engineering data were also smaller than in typical force fields and followed the “like dissolves like” rule, as predicted by McLachlan theory .Popular force fields
Different force fields are designed for different purposes.
MM2 was developed primarily for conformational analysis of small organic molecules. It is designed to reproduce the equilibrium covalent geometry of molecules as precisely as possible. It implements a large set of parameters that is continuously refined and updated for many different classes of organic compounds (MM3 andMM4 ).CFF was developed by Warshel, Lifson and coworkers as a general method for unifying studies of energies, structures and vibration of general molecules and molecular crystals. The CFF program, developed by Levitt and Warshel, is based on the Cartesian representation of all the atoms, and it served as the basis for many subsequent simulation programs.
ECEPP was developed specifically for modeling of peptides and proteins. It uses fixed geometries of amino acid residues to simplify the potential energy surface. Thus, the energy minimization is conducted in the space of protein torsion angles. Both
MM2 and ECEPP include potentials for H-bonds and torsion potentials for describing rotations around single bonds. ECEPP/3 was implemented (with some modifications) inInternal Coordinate Mechanics and FANTOM Schaumann, T., Braun, W. and Wutrich, K. (1990) The program FANTOM for energy refinement of polypeptides and proteins using a Newton-Raphson minimizer in torsion angle space. "Biopolymers" 29: 679-694.] .AMBER ,CHARMM andGROMOS have been developed primarily formolecular dynamics of macromolecules, although they are also commonly applied for energy minimization. Therefore, the coordinates of all atoms are considered as free variables.Classical force fields
* [http://amber.scripps.edu/ AMBER (Assisted Model Building and Energy Refinement)] - widely used for proteins and DNA
* [http://www.charmm.org/ CHARMM (Chemistry at HARvard Molecular Mechanics)] - originally developed at Harvard, widely used for both small molecules and macromolecules
*CHARMm - commercial version of CHARMM, available throughAccelrys
*CVFF - also broadly used for small molecules and macromolecules
*GROMACS - The force field optimized for the package of the same name
*GROMOS - A force field that comes as part of the [http://www.igc.ethz.ch/gromos/ GROMOS] (GROningen MOlecular Simulation package), a general-purpose molecular dynamics computer simulation package for the study of biomolecular systems. GROMOS force field (A-version) has been developed for application to aqueous or apolar solutions of proteins, nucleotides and sugars. However, a gas phase version (B-version) for simulation of isolated molecules is also available
*OPLS (Optimized Potential for Liquid Simulations) (variations include OPLS-AA, OPLS-UA, OPLS-2001, OPLS-2005) developed by [http://zarbi.chem.yale.edu/ William L. Jorgensen] at the Yale University Department of Chemistry.
* [http://laetro.usc.edu/programs/#molaris ENZYMIX] – A general polarizable force field for modeling chemical reactions in biological molecules. This force field is implemented with the empirical valence bond (EVB) method and is also combined with the semimacroscopic PDLD approach in the program in the [http://futura.usc.edu/programs/index.html#molaris MOLARIS] package.
* [http://msdlocal.ebi.ac.uk/docs/mmrefs.html#ecepp ECEPP/2] - First force field for polypeptide molecules - developed by F.A.Momany, H.A.Scheraga and colleagues.
*QCFF/PI – A general force field for conjugated molecules. Warshel A (1973). Quantum Mechanical Consistent Force Field (QCFF/PI) Method: Calculations of Energies, Conformations and Vibronic Interactions of Ground and Excited States of Conjugated Molecules, Israel J. Chem. 11: 709.] . D.W.M. Hofmann, L. N. Kuleshova, B. D'Aguanno (2007) A new reactive potential for the Molecular Dynamics simulation of liquid water. Chem.Phys.Lett. 448: 138-143]Other
*
VALBOND - a function for angle bending that is based onvalence bond theory and works for large angular distortions,hypervalent molecule s, and transition metal complexes. It can be incorporated into other force fields such as CHARMM and UFF.Water Models
"Main article:
water model The set of parameters used to model water or aqueous solutions (basically a force field for water) is called a
water model . Water has attracted a great deal of attention due to its unusual properties and its importance as a solvent. Many water models have been proposed; some examples are TIP3P, TIP4P, SPC, and ST2.See also
*
Molecular dynamics
*Molecular modelling
* [http://www.agilemolecule.com/Manual/Construction.html Molecular model construction with proper Force field association.] [http://www.agilemolecule.com/AGM_Build.html (AGM Build)]
*Software for molecular mechanics modeling References
Further reading
# Schlick T. (2000). "Molecular Modeling and Simulation: An Interdisciplinary Guide" Interdisciplinary Applied Mathematics: Mathematical Biology. Springer-Verlag New York, NY.
# Israelachvili, J.N. (1992) "Intermolecular and surface forces." Academic Press, San Diego.
#Warshel A (1991). "Computer Modeling of Chemical Reactions in Enzymes and Solutions" John Wiley & Sons New York.
Wikimedia Foundation. 2010.