- Saposin protein domain
-
Identifiers Symbol SapA Pfam PF02199 InterPro IPR003119 PROSITE PDOC51110 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Saposin-like type B, region 1 
Crystal structure of human saposin C dimer in an open conformation.[1] Identifiers Symbol SapB_1 Pfam PF05184 InterPro IPR007856 PROSITE PDOC50015 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Identifiers Symbol SapB_2 Pfam PF03489 InterPro IPR008138 PROSITE PDOC50015 SCOP 1nkl OPM family 83 OPM protein 1sn6 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Saposins are small lysosomal proteins that serve as activators of various lysosomal lipid-degrading enzymes.[2] They probably act by isolating the lipid substrate from the membrane surroundings, thus making it more accessible to the soluble degradative enzymes. All mammalian saposins are synthesized as a single precursor molecule (prosaposin) which contains four Saposin-B domains, yielding the active saposins after proteolytic cleavage, and two Saposin-A domains that are removed in the activation reaction. The Saposin-B domains also occur in other proteins, many of them active in the lysis of membranes.[3][4]
Contents
Domain organization
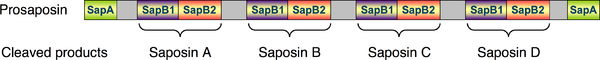
Below is a schematic diagram of the 1D structure of the prosaposin protein depicting the N- and C-terminal SapA domains and the four SapB1 and four SapB2 domains. Proteolytic cleavage of the proprotein occurs in the grey regions. Adjacent pairs of SapB1 and SapB2 domains remain connected after proteolytic processing of prosaposin and each pair comprises one of the mature saponin A-D proteins.
Human proteins containing this domain
- AOAH
- GNLY
- Prosaposin
- PSAPL1
- SFTPB
References
- ^ PDB 2qyp, Rossmann M, Schultz-Heienbrok R, Behlke J, Remmel N, Alings C, Sandhoff K, Saenger W, Maier T (May 2008). "Crystal structures of human saposins C and D: implications for lipid recognition and membrane interactions". Structure 16 (5): 809–17. doi:10.1016/j.str.2008.02.016. PMID 18462685.
- ^ Munford RS, Sheppard PO, O Hara PJ (1995). "Saposin-like proteins (SAPLIP) carry out diverse functions on a common backbone structure". J. Lipid Res. 36 (8): 1653–1663. PMID 7595087.
- ^ Ponting CP (1994). "Acid sphingomyelinase possesses a domain homologous to its activator proteins: saposins B and D". Protein Sci. 3 (2): 359–361. doi:10.1002/pro.5560030219. PMC 2142785. PMID 8003971. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2142785.
- ^ Hofmann K, Tschopp J (1996). "Cytotoxic T cells: more weapons for new targets?". Trends Microbiol. 4 (3): 91–94. doi:10.1016/0966-842X(96)81522-8. PMID 8868085.
Further reading
- Ponting CP, Russell RB (May 1995). "Swaposins: circular permutations within genes encoding saposin homologues". Trends in Biochemical Sciences 20 (5): 179–80. doi:10.1016/S0968-0004(00)89003-9. PMID 7610480.
External links

This membrane protein-related article is a stub. You can help Wikipedia by expanding it.
