- Cladogram
-
For help on how to use cladograms in Wikipedia, see Help:Cladograms
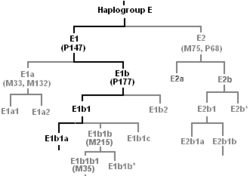
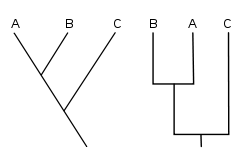
A cladogram is a diagram used in cladistics which shows ancestral relations between organisms, to represent the evolutionary tree of life. Although traditionally such cladograms were generated largely on the basis of morphological characters, DNA and RNA sequencing data and computational phylogenetics are now very commonly used in the generation of cladograms.
Contents
Generating a cladogram
A greatly simplified procedure for generating a cladogram is:[1]
- Gather and organize data
- Consider possible cladograms
- Select best cladogram
Step 1: gather and organize data
A cladistic analysis begins with the following data:
- a list of taxa (for example, species) to be organized
- a list of characteristics to be compared
- for each taxon, the value of each of the listed characteristics or character states
For example, if analyzing 20 species of birds, the data might be:
- the list of the 20 species
- characteristics such as genome sequence, skeletal anatomy, biochemical processes, and feather coloration
- for each of the 20 species, its particular genome sequence, skeletal anatomy, biochemical processes, and feather coloration
All the data are then organized into a "taxon-character matrix", which is the base to perform phylogenetic analysis.
Molecular versus morphological data
The characteristics used to create a cladogram can be roughly categorized as either morphological (synapsid skull, warm blooded, notochord, unicellular, etc.) or molecular (DNA, RNA, or other genetic information).[1] Prior to the advent of DNA sequencing, all cladistic analysis used morphological data.
As DNA sequencing has become cheaper and easier, molecular systematics has become a more and more popular way to reconstruct phylogenies.[2] Using a parsimony criterion is only one of several methods to infer a phylogeny from molecular data; maximum likelihood and Bayesian inference, which incorporate explicit models of sequence evolution, are non-Hennigian ways to evaluate sequence data. Another powerful method of reconstructing phylogenies is the use of genomic retrotransposon markers, which are thought to be less prone to the problem of reversion that plagues sequence data. They are also generally assumed to have a low incidence of homoplasies because it was once thought that their integration into the genome was entirely random; this seems at least sometimes not to be the case, however.
Ideally, morphological, molecular, and possibly other phylogenies should be combined into an analysis of total evidence: All have different intrinsic sources of error. For example, character convergence (homoplasy) is much more common in morphological data than in molecular sequence data, but character reversions that are unrecognizable as such are more common in the latter (see long branch attraction). Morphological homoplasies can usually be recognized as such if character states are defined with enough attention to detail.
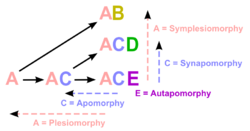
Plesiomorphies and synapomorphies
The researcher must decide which character states were present before the last common ancestor of the species group (plesiomorphies) and which were present in the last common ancestor (synapomorphies) and does so by comparison to one or more outgroups. The choice of an outgroup is a crucial step in cladistic analysis because different outgroups can produce trees with profoundly different topologies. Note that only synapomorphies are of use in characterizing clades.[citation needed]
Avoid homoplasies
A homoplasy is a character that is shared by multiple species due to some cause other than common ancestry.[3] The two main types of homoplasy are convergence (appearance of the same character in at least two distinct lineages) and reversion (the return to an ancestral character). Use of homoplasies when building a cladogram is sometimes unavoidable but is to be avoided when possible.
A well known example of homoplasy due to convergent evolution would be the character, "presence of wings". Though the wings of birds, bats, and insects serve the same function, each evolved independently, as can be seen by their anatomy. If a bird, bat, and a winged insect were scored for the character, "presence of wings", a homoplasy would be introduced into the dataset, and this would confound the analysis, possibly resulting in a false evolutionary scenario.
Homoplasies can often be avoided outright in morphological datasets by defining characters more precisely and increasing their number. When analyzing "supertrees" (datasets incorporating as many taxa of a suspected clade as possible), it may become unavoidable to introduce character definitions that are imprecise, as otherwise the characters might not apply at all to a large number of taxa; to continue with the "wings" example, the presence of wings would hardly be a useful character if attempting a phylogeny of all Metazoa, as most of these don't have wings at all. Cautious choice and definition of characters thus is another important element in cladistic analyses. With a faulty outgroup or character set, no method of evaluation is likely to produce a phylogeny representing the evolutionary reality.
Step 2: consider possible cladograms
Main article: Computational phylogeneticsWhen there are just a few species being organized, it is possible to do this step manually, but most cases require a computer program. There are scores of computer programs available to support cladistics.[4] See phylogenetic tree for more information about tree-generating computer programs.
Because the total number of possible cladograms grows factorially[5] with the number of species, it is impractical for a computer program to evaluate every individual cladogram. A typical cladistic program begins by using heuristic techniques to identify a small number of candidate cladograms. Many cladistic programs then continue the search with the following repetitive steps:
- Evaluate the candidate cladograms by comparing them to the characteristic data
- Identify the best candidates that are most consistent with the characteristic data
- Create additional candidates by creating several variants of each of the best candidates from the prior step
- Use heuristics to create several new candidate cladograms unrelated to the prior candidates
- Repeat these steps until the cladograms stop getting better
Computer programs that generate cladograms use algorithms that are very computationally intensive,[6] because the cladogram problem is NP-hard.
Step 3: select best cladogram
There are several algorithms available to identify the "best" cladogram.[7] Most algorithms use a metric to measure how consistent a candidate cladogram is with the data. Most cladogram algorithms use the mathematical techniques of optimization and minimization.
In general, cladogram generation algorithms must be implemented as computer programs, although some algorithms can be performed manually when the data sets are trivial (for example, just a few species and a couple of characteristics).
Some algorithms are useful only when the characteristic data are molecular (DNA, RNA); other algorithms are useful only when the characteristic data are morphological. Other algorithms can be used when the characteristic data includes both molecular and morphological data.
Algorithms for cladograms include least squares, neighbor-joining, parsimony, maximum likelihood, and Bayesian inference.
Biologists sometimes use the term parsimony for a specific kind of cladogram generation algorithm and sometimes as an umbrella term for all cladogram algorithms.[8]
Algorithms that perform optimization tasks (such as building cladograms) can be sensitive to the order in which the input data (the list of species and their characteristics) is presented. Inputting the data in various orders can cause the same algorithm to produce different "best" cladograms. In these situations, the user should input the data in various orders and compare the results.
Using different algorithms on a single data set can sometimes yield different "best" cladograms, because each algorithm may have a unique definition of what is "best".
Because of the astronomical number of possible cladograms, algorithms cannot guarantee that the solution is the overall best solution. A nonoptimal cladogram will be selected if the program settles on a local minimum rather than the desired global minimum.[9] To help solve this problem, many cladogram algorithms use a simulated annealing approach to increase the likelihood that the selected cladogram is the optimal one.[10]
Cladogram Statistics
Consistency index
The consistency index (CI) measures the amount of homoplasy in a cladogram. It is calculated by counting the mininum number of changes in a dataset and dividing it by the actual number of changes required on the cladogram.
Retention index
The retention index (RI) is also a measure of the amount of homoplasy but also measures how well synapomorphies explain the tree. It is calculated taking the product of the maximum number of changes on a tree and the number of changes on the tree divided by the product of the maximum number of changes on the tree and the minimum number of changes in the dataset.
The rescaled retention index (RC) is obtained by multiplying the CI by the RI. The homoplasy index (HI) is simply 1-CI.
Incongruence length difference test (or partition homogeneity test)
The incongruence length difference test (ILD) is a measurement of how the combination of different datasets (e.g. morphological and molecular, plastid and nuclear genes) contributes to a longer tree. It is measured by first calculating the total tree length of each partition and summing them. Then replicates are made by making randomly assembled partitions consisting of the original partitions. The lengths are summed. A p value of 0.01 is obtained for 100 replicates if 99 replicates have longer combined tree lengths.
References
- ^ a b DeSalle, Rob (2002). Techniques in Molecular Systematics and Evolution. Birkhauser. ISBN 376436257X.
- ^ Hillis, David (1996). Molecular Systematics. Sinaur. ISBN 0878932828.
- ^ West-Eberhard, Mary Jane (2003). Developmental Plasticity and Evolution. Oxford Univ. Press. pp. 353–376. ISBN 0195122356.
- ^ "List of Cladistics Software Programs". http://evolution.genetics.washington.edu/phylip/software.pars.html.
- ^ Felsenstein, Joseph (2004). Inferring Phylogenies. Sunderland, MA: Sinauer Associates, Inc.. pp. 23. ISBN 0-87893-177-5.
- ^ Hodkinson, Trevor (2006). Reconstructing the Tree of Life: Taxonomy and Systematics of Species Rich Taxa. CRC Press. p. 61–128. ISBN 0849395798.
- ^ Kitching, Ian (1998). Cladistics: The Theory and Practice of Parsimony Analysis. Oxford University Press. ISBN 0198501382.
- ^ Stewart, Caro-Beth (1993). "The Powers and Pitfalls of Parsimony". Nature 361 (6413): 603–607. doi:10.1038/361603a0. PMID 8437621.
- ^ Foley, Peter (1993). Cladistics: A Practical Course in Systematics. Oxford Univ. Press. p. 66. ISBN 0198577664.
- ^ Nixon K. C. (1999). "The Parsimony Ratchet: a new method for rapid parsimony analysis". Cladistics 15 (4): 407–414. doi:10.1111/j.1096-0031.1999.tb00277.x.
Categories:- Diagrams
- Phylogenetics
Wikimedia Foundation. 2010.