- Transmembrane protein
A transmembrane protein is a
protein that spans the entirebiological membrane . Transmembrane proteins aggregate and precipitate in water. They requiredetergent s or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
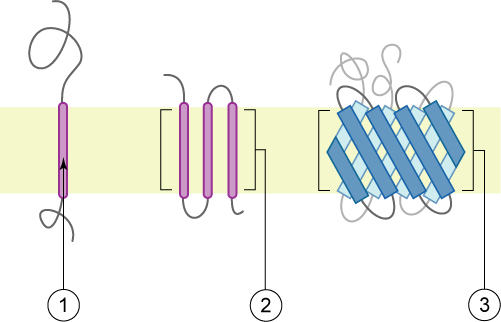
thumb|right|400px|Schematic representation of transmembrane proteins: 1. a single transmembrane α-helix (bitopic membrane protein)2. a polytopic α-helical protein3. a transmembrane β barrel ">
The membrane is represented in light brown.Types
There are two basic types of transmembrane proteins:
#Alpha-helical. These proteins are present in the inner membranes of bacterial cells or the plasma membrane of eukaryotes, and sometimes in theouter membrane s [alpha-helical proteins in outer membranes includeStannin and [http://opm.phar.umich.edu/protein.php?search=2j58 certain lipoproteins] , and others] This is the major category of transmembrane proteins.
#Beta-barrels. These proteins are so far found only inouter membrane s ofGram-negative bacteria ,cell wall ofGram-positive bacteria , andouter membrane s ofmitochondria andchloroplasts . All beta-barrel transmembrane proteins have simplest up-and-down topology, which may reflect their common evolutionary origin and similar folding mechanism.Thermodynamic stability and folding
tability of α-helical transmembrane proteins
Transmembrane α-helical proteins are unusually stable judging from thermal denaturation studies, because they do not unfold completely within the membranes (the complete unfolding would require breaking down too many α-helical
H-bond s in the nonpolar media). On the other hand, these proteins easily "misfold", due to non-native aggregation in membranes, transition to themolten globule states, formation of non-nativedisulfide bonds , or unfolding of peripheral regions and nonregular loops that are locally less stable.It is also important to properly define the "
unfolded state ". The "unfolded state" of membrane proteins indetergent micelles is different from that in the thermal denaturation experiments. This state represents a combination of folded hydrophobic α-helices and partially unfolded segments covered by the detergent. For example, the "unfolded"bacteriorhodopsin in SDS micelles has four transmembrane α-helices folded, while the rest of the protein is situated at the micelle-water interface and can adopt different types of non-nativeamphiphilic structures. Free energy differences between such detergent-denatured and native states are similar to stabilities of water-soluble proteins (< 10 kcal/mol).Folding of α-helical transmembrane proteins
Refolding of α-helical transmembrane proteins "in vitro" is technically difficult. There are relatively few examples of the successful refolding experiments, as for
bacteriorhodopsin . "In vivo" all such proteins are normally folded co-translationally within the large transmembranetranslocon . The translocon channel provides a highly heterogeneous environment for the nascent transmembane α-helices. A relatively polar amphiphilic α-helix can adopt a transmembrane orientation in the translocon (although it would be at the membrane surface or unfolded "in vitro"), because its polar residues can face the central water-filled channel of the translocon. Such mechanism is necessary for incorporation of polar α-helices into structures of transmembrane proteins. The amphiphilic helices remain attached to the translocon until the protein is completely synthesized and folded. If the protein remains unfolded and attached to the translocon for too long, it is degraded by specific "quality control" cellular systems.tability and folding of β-barrel transmembrane proteins
Stability of β-barrel transmembrane proteins is similar to stability of water-soluble proteins, based on chemical denaturation studies. Their folding "in vivo" is facilitated by water-soluble chaperones, such as protein Skp [http://faculty.virginia.edu/tamm/pages/BBA_Folding_Review.pdf] .
3D structures
Light absorption-driven transporters
*
Bacteriorhodopsin -like proteins includingrhodopsin (see alsoopsin ) [http://opm.phar.umich.edu/families.php?superfamily=6]
*Bacterialphotosynthetic reaction centre s andphotosystem s I and II [http://opm.phar.umich.edu/families.php?superfamily=2]
*Light harvesting complex es frombacteria andchloroplasts [http://opm.phar.umich.edu/families.php?superfamily=1]Oxidoreduction-driven transporters
*Transmembrane cytochrome b-like proteins [http://opm.phar.umich.edu/families.php?superfamily=3] :
coenzyme Q - cytochrome c reductase (cytochrome bc1 );cytochrome b6f complex ; formate dehydrogenase, respiratorynitrate reductase ;succinate - coenzyme Q reductase (fumarate reductase); andsuccinate dehydrogenase . Seeelectron transport chain .
*Cytochrome c oxidase s [http://opm.phar.umich.edu/families.php?superfamily=4] frombacteria andmitochondria Electrochemical potential-driven transporters
*Proton or sodium translocating F-type and V-type
ATPase s [http://opm.phar.umich.edu/families.php?superfamily=5]P-P-bond hydrolysis-driven transporters
*P-type
calcium ATPase (five different conformations) [http://opm.phar.umich.edu/families.php?superfamily=22]
*Calcium ATPase regulatorsphospholamban and sarcolipin [http://opm.phar.umich.edu/families.php?superfamily=70]
*ABC transporter s: [http://opm.phar.umich.edu/families.php?superfamily=17 BtuCD] , [http://opm.phar.umich.edu/families.php?superfamily=18 multidrug transporter] , and [http://opm.phar.umich.edu/families.php?superfamily=198 molybdate uptake transporter]
*Generalsecretory pathway (Sec)translocon (preprotein translocase SecY) [http://opm.phar.umich.edu/families.php?superfamily=19]Porters (uniporters,
symporters ,antiporters )*
Mitochondrial carrier protein s [http://opm.phar.umich.edu/families.php?superfamily=21]
*Major Facilitator Superfamily (Glycerol-3-hosphate transporter, Lactosepermease , and Multidrug transporter EmrD) [http://opm.phar.umich.edu/families.php?superfamily=15]
*Resistance-nodulation-cell division (multidrugefflux transporter AcrB, seemultidrug resistance ) [http://opm.phar.umich.edu/families.php?superfamily=16]
*Dicarboxylate/amino acid:cation symporter (proton glutamate symporter) [http://opm.phar.umich.edu/families.php?superfamily=20]
*Monovalent cation/proton antiporter (Sodium/proton antiporter 1 NhaA) [http://opm.phar.umich.edu/families.php?superfamily=66]
*Neurotransmitter sodium symporter [http://opm.phar.umich.edu/families.php?superfamily=67]
*Ammonia transporters [http://opm.phar.umich.edu/families.php?superfamily=13]
*Drug/Metabolite Transporter (small multidrug resistance transporter EmrE - the structures are retracted as erroneous) [http://opm.phar.umich.edu/families.php?superfamily=77]Alpha-helical channels including
ion channels *
Voltage-gated ion channel like, includingpotassium channel s KcsA and KvAP, andinward-rectifier potassium ion channel Kirbac [http://opm.phar.umich.edu/families.php?superfamily=8]
*Large-conductance mechanosensitive channel, MscL [http://opm.phar.umich.edu/families.php?superfamily=12]
*Small-conductance mechanosensitive ion channel (MscS) [http://opm.phar.umich.edu/families.php?superfamily=11]
*CorA metal ion transporters [http://opm.phar.umich.edu/families.php?superfamily=72]
*Ligand-gated ion channel ofneurotransmitter receptors (acetylcholine receptor ) [http://opm.phar.umich.edu/families.php?superfamily=14]
*Aquaporin s [http://opm.phar.umich.edu/families.php?superfamily=7]
*Chloride channel s [http://opm.phar.umich.edu/families.php?superfamily=10]
*Outer membrane auxiliary proteins (polysaccharide transporter) [http://opm.phar.umich.edu/families.php?superfamily=188] - α-helical transmembrane proteins from the outer bacterial membraneEnzymes
*
Methane monooxygenase [http://opm.phar.umich.edu/families.php?superfamily=23]
*Rhomboid protease [http://opm.phar.umich.edu/families.php?superfamily=186]
*Disulfide bond formation protein (DsbA-DsbB complex) [http://opm.phar.umich.edu/protein.php?pdbid=2hi7]Proteins with alpha-helical transmembrane anchors
*
T cell receptor transmembrane dimerization domain [http://opm.phar.umich.edu/families.php?superfamily=187]
*Cytochrome cnitrite reductase complex [http://opm.phar.umich.edu/protein.php?pdbid=2j7a]
*Steryl-sulfate sulfohydrolase [http://opm.phar.umich.edu/families.php?superfamily=24]
*Stannin [http://opm.phar.umich.edu/families.php?superfamily=180]
*Glycophorin A dimer [http://opm.phar.umich.edu/families.php?superfamily=25]
*Inovirus (filamentous phage ) major coat protein [http://opm.phar.umich.edu/families.php?superfamily=73]
*Pilin [http://opm.phar.umich.edu/families.php?superfamily=74]
*Pulmonary surfactant -associated protein [http://opm.phar.umich.edu/families.php?superfamily=75]
*Monoamine oxidase s A and B [http://opm.phar.umich.edu/families.php?family=176] ,
*Cytochrome P450 oxidase s [http://opm.phar.umich.edu/families.php?superfamily=41] ,
*Corticosteroid 11β-dehydrogenases [http://opm.phar.umich.edu/families.php?superfamily=127] .
*Signal Peptide Peptidase [http://opm.phar.umich.edu/families.php?superfamily=178]
*Membrane protease specific for a stomatin homolog [http://opm.phar.umich.edu/protein.php?pdbid=2deo]β-barrels composed of a single polypeptide chain
*Beta barrels from eight beta-strands and with "shear number" of ten ("n=8, S=10") [http://opm.phar.umich.edu/families.php?superfamily=26] . They include:
**OmpA-like transmembrane domain (OmpA),
**Virulence-related outer membrane protein family (OmpX),
**Outer membrane protein W family (OmpW),
**Antimicrobial peptide resistance and lipid A acylation protein family (PagP)
**Lipid A deacylase PagL , and
**Opacity family porins (NspA)
*Autotransporter domain ("n=12,S=14') [http://opm.phar.umich.edu/families.php?superfamily=28]
*FadL outer membrane protein transport family , includingFatty acid transporter FadL ("n=14,S=14") [http://opm.phar.umich.edu/families.php?superfamily=30]
*General bacterial porin family , known as trimeric porins ("n=16,S=20") [http://opm.phar.umich.edu/families.php?superfamily=31]
*Maltoporin , or sugar porins ("n=18,S=22") [http://opm.phar.umich.edu/families.php?superfamily=32]
*Nucleoside-specific porin ("n=12,S=16") [http://opm.phar.umich.edu/families.php?superfamily=71]
*Outer membrane phospholipase A1 ("n=12,S=16") [http://opm.phar.umich.edu/families.php?superfamily=29]
*TonB-dependent receptors and their plug domain. They are ligand-gated outer membrane channels ("n=22,S=24"), includingcobalamin transporter BtuB, Fe(III)-pyochelin receptor FptA, receptor FepA, ferric hydroxamate uptake receptor FhuA, transporter FecA, and pyoverdine receptor FpvA [http://opm.phar.umich.edu/families.php?superfamily=33]
*Outer membrane protein OpcA family ("n=10,S=12") that includes outer membraneprotease OmpT andadhesin /invasin OpcA protein [http://opm.phar.umich.edu/families.php?superfamily=27]
*Outer membrane protein G porin family ("n=14,S=16") [http://opm.phar.umich.edu/families.php?superfamily=181]Note: "n" and "S" are, respectively, the number of beta-strands and the "shear number" [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=8126726&query_hl=3&itool=pubmed_docsum] of the
beta-barrel β-barrels composed of several polypeptide chains
*Trimeric autotransporter ("n=12,S=12") [http://opm.phar.umich.edu/families.php?superfamily=179]
*Outer membrane efflux proteins , also known as trimeric outer membrane factors (n=12,S=18) including TolC and multidrug resistance proteins [http://opm.phar.umich.edu/families.php?superfamily=34]
*MspA porin (octamer, "n=S=16") and α-hemolysin (heptamer "n=S=14") [http://opm.phar.umich.edu/families.php?superfamily=35] . These proteins are secreted.See also
Gramicidin A [http://opm.phar.umich.edu/protein.php?pdbid=1grm] , a peptide that forms a dimeric transmembrane β-helix. It is also secreted byGram-positive bacteria.References
*Booth, P.J., Templer, R.H., Meijberg, W., Allen, S.J., Curran, A.R., and Lorch, M. 2001. In vitro studies of membrane protein folding. "Crit. Rev. Biochem. Mol. Biol." 36: 501-603.
*Bowie J.U. 2001. Stabilizing membrane proteins. "Curr. Op. Struct. Biol." 11: 397-402.
*Bowie J.U. 2005. Solving the membrane protein folding problem. "Nature" 438: 581-589.
*DeGrado W.F., Gratkowski H. and Lear J.D. 2003. How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. "Protein Sci." 12: 647-665.
*Lee, A.G. 2003 Lipid-protein interactions in biological membranes: a structural perspective. "Biochim. Biophys. Acta" 1612: 1-40.
*Lee, A.G. 2004. How lipids affect the activities of integral membrane proteins. "Biochim. Biophys. Acta" 1666: 62-87.
*le Maire, M., Champeil, P., and Moller, J.V. 2000. Interaction of membrane proteins and lipids with solubilizing detergents. "Biochim. Biophys. Acta" 1508: 86-111.
*Popot J-L. and Engelman D.M. 2000. Helical membrane protein folding, stability, and evolution. "Annu. Rev. Biochem." 69: 881-922.
*"Protein-lipid interactions" (Ed. L.K. Tamm) Wiley, 2005.
*Tamm, L.K., Hong, H., and Liang, B.Y. 2004. Folding and assembly of beta-barrel membrane proteins. "Biochim. Biophys. Acta" 1666: 250-263.Additional examples
*Some
cell adhesion proteins
*Some receptor proteins
*Insulin receptor
*GLUTI
*Integrin
*Cadherin External links
* [http://www.tcdb.org/ TCDB] - Transporter classification database from Milton H. Saier, Jr. laboratory
* [http://www.membranetransport.org/ TransportDB] Genomics-oriented database of transporters from TIGR
* [http://www.mpdb.ul.ie/ Membrane PDB] Database of 3D structures of integral membrane proteins and hydrophobic peptides with an emphasis on crystallization conditions
* [http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html Membrane proteins of known 3D structure] from Stephen White laboratory
* [http://pdbtm.enzim.hu/ PDBTM] "All" 3D models of transmembrane peptides and proteins currently in the PDB including theoretical models. Approximate positions of membrane boundary planes were calculated for each PDB entry.
* [http://opm.phar.umich.edu/ Orientations of proteins in membranes database] - Calculated spatial positions of transmembrane, integral monotopic, and peripheral proteins in membranesee also
*
cell membrane
*transmembrane receptor s
*membrane topology
*transmembrane helix
*membrane protein
*integral membrane protein
*peripheral membrane protein Transporter Classification database
Wikimedia Foundation. 2010.