- Gram-positive bacteria
-
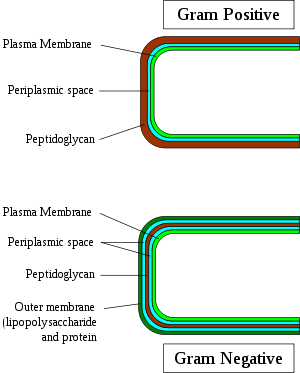
Gram-positive bacteria are those that are stained dark blue or violet by Gram staining. This is in contrast to Gram-negative bacteria, which cannot retain the crystal violet stain, instead taking up the counterstain (safranin or fuchsine) and appearing red or pink. Gram-positive organisms are able to retain the crystal violet stain because of the high amount of peptidoglycan in the cell wall. Gram-positive cell walls typically lack the outer membrane found in Gram-negative bacteria.
Contents
Characteristics
The following characteristics are generally present in a Gram-positive bacterium:[1]
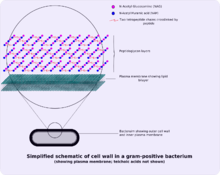
- cytoplasmic lipid membrane
- thick peptidoglycan layer
- teichoic acids and lipoids are present, forming lipoteichoic acids, which serve to act as chelating agents, and also for certain types of adherence.
- capsule polysaccharides (only in some species)
- flagellum (only in some species)
- if present, it contains two rings for support as opposed to four in Gram-negative bacteria because Gram-positive bacteria have only one membrane layer.
- The individual peptidoglycan molecules are cross-linked by pentaglycine chains by a DD-transpeptidase enzyme. In gram-negative bacteria, the transpeptidase creates a covalent bond directly between peptidoglycan molecules, with no intervening bridge.
Both Gram-positive and Gram-negative bacteria may have a membrane called an S-layer. In Gram-negative bacteria, the S-layer is attached directly to the outer membrane. In Gram-positive bacteria, the S-layer is attached to the peptidoglycan layer. Unique to Gram-positive bacteria is the presence of teichoic acids in the cell wall. Some particular teichoic acids, lipoteichoic acids, have a lipid component and can assist in anchoring peptidoglycan, as the lipid component is embedded in the membrane.
Classification
Along with cell shape, Gram staining is a rapid diagnostic tool of use to group species of Bacteria. In traditional and even some areas of contemporary microbiological practice, such staining, alongside growth requirement and antibiotic susceptibility testing, and other macroscopic and physiologic tests, forms the full basis for classification and subdivision of the Bacteria (e.g., see Figure, and pre-1990 versions of Bergey's Manual).
As such, historically, the kindgom Monera was divided into four divisions based primarily on Gram staining: Firmicutes (positive in staining), Gracillicutes (negative in staining), Mollicutes (neutral in staining) and Mendocutes (variable in staining).[2]
Since 1987 and the seminal 16S ribosomal RNA phylogenetic studies of Carl Woese (Department of Microbiology, University of Illinois) and collaborators and colleagues, the monophyly of the Gram-positive bacteria has been challenged,[3] with striking productive implications for the therapeutic and general study of these organisms. Based on molecular studies of 16S sequences, Woese recognised 12 bacterial phyla, two being Gram-positive: high-GC Gram-positives and low-GC Gram-positives (where G and C refer to the guanine and cytosine content in their genomes),[3] which are now referred to by these names, or as Actinobacteria and Firmicutes. The former, the Actinobacteria, are the high GC content Gram-positive bacteria. The latter, the Firmicutes are the "low-GC" Gram-positive bacteria, which actually have 45%–60% GC content but lower than that of the Actinobacteria,[1]. The Firmicutes contain the well-known genera that are majority of Gram-positives of medical interest: Staphylococcus, Streptococcus, Enterococcus (cocci), Bacillus, Corynebacterium, Nocardia, Clostridium, Actinobacteria and Listeria (bacilli/rods). This group also been expanded to include the Mycoplasma, or Mollicutes, bacteria-like organisms that lack cell walls and cannot be Gram-stained, but appear to have derived evolutionarily from such forms.
Despite the wide acceptance and practical record of utility of the new molecular phylogeny, a small group, including Cavalier-Smith, still treat the Monera as a monophyletic clade and refer to the group as division "Posibacteria".[4]
Exceptions
In general, Gram-positive bacteria have a single lipid bilayer (monoderms), whereas Gram-negative have two (diderms). Some taxa lack peptidoglycan (such as the domain Archaea, the class Mollicutes, some members of the Rhickettsiales, and the insect-endosymbionts of the Enterobacteriales) and are Gram-variable. This, however, does not always hold true.
The Deinococcus-Thermus bacteria have Gram-positive stains, although they are structurally similar to Gram-negative bacteria with two layers (diderms).
The Chloroflexi have a single layer, yet (with some exceptions[5]) stain negative.[6] Two related phyla to the Chloroflexi, the TM7 clade and the Ktedonobacteria, are also monoderms.[7][8]
Some Firmicute species are not Gram-positive; these belong to the class Mollicutes (alternatively considered a class of the phylum Tenericutes), which lack peptidoglycan (Gram-indeterminate), and the class Negativicutes, which includes Selenomonas and which stain Gram-negative.[9]
Pathogenesis
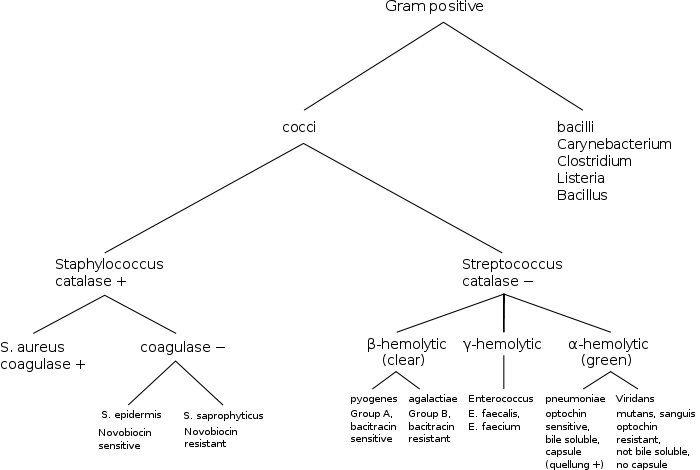
Most pathogens in humans are Gram-positive organisms. In the classical sense, six Gram-positive genera are typically pathogenic in humans. Two of these, Streptococcus and Staphylococcus, are cocci (sphere-shaped bacteria). The remaining organisms are bacilli (rod-shaped bacteria) and can be subdivided based on their ability to form spores. The non-spore formers are Corynebacterium and Listeria (a coccobacillus), whereas Bacillus and Clostridium produce spores.[10] The spore-forming bacteria can again be divided based on their respiration: Bacillus is a facultative anaerobe, while Clostridium is an obligate anaerobe.
See also
- Gram-negative
- Gram staining
References
- ^ a b Madigan M; Martinko J (editors). (2005). Brock Biology of Microorganisms (11th ed.). Prentice Hall. ISBN 0131443291.
- ^ Gibbons, N. E.; Murray, R. G. E. (1978). "Proposals Concerning the Higher Taxa of Bacteria". IJSEM 28 (1): 1–6. doi:10.1099/00207713-28-1-1.
- ^ a b Woese, CR (1987). "Bacterial evolution". Microbiological reviews 51 (2): 221–71. PMC 373105. PMID 2439888. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=373105.
- ^ Cavalier-Smith T (2006). "Rooting the tree of life by transition analyses". Biol. Direct 1: 19. doi:10.1186/1745-6150-1-19. PMC 1586193. PMID 16834776. http://www.biology-direct.com/content/1//19.
- ^ Yabe, S.; Aiba, Y.; Sakai, Y.; Hazaka, M.; Yokota, A. (2010). "Thermogemmatispora onikobensis gen. nov., sp. nov. And Thermogemmatispora foliorum sp. nov., isolated from fallen leaves on geothermal soils, and description of Thermogemmatisporaceae fam. Nov. And Thermogemmatisporales ord. Nov. Within the class Ktedonobacteria". International Journal of Systematic and Evolutionary Microbiology 61 (4): 903–910. doi:10.1099/ijs.0.024877-0. PMID 20495028.
- ^ Sutcliffe, I. C. (2011). "Cell envelope architecture in the Chloroflexi: A shifting frontline in a phylogenetic turf war". Environmental Microbiology 13 (2): 279–282. doi:10.1111/j.1462-2920.2010.02339.x. PMID 20860732.
- ^ Hugenholtz, P.; Tyson, G. W.; Webb, R. I.; Wagner, A. M.; Blackall, L. L. (2001). "Investigation of Candidate Division TM7, a Recently Recognized Major Lineage of the Domain Bacteria with No Known Pure-Culture Representatives". Applied and Environmental Microbiology 67 (1): 411. doi:10.1128/AEM.67.1.411-419.2001. PMC 92593. PMID 11133473. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=92593.
- ^ Cavaletti, L.; Monciardini, P.; Bamonte, R.; Schumann, P.; Rohde, M.; Sosio, M.; Donadio, S. (2006). "New Lineage of Filamentous, Spore-Forming, Gram-Positive Bacteria from Soil". Applied and Environmental Microbiology 72 (6): 4360–4369. doi:10.1128/AEM.00132-06. PMC 1489649. PMID 16751552. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1489649.
- ^ Marchandin, H.; Teyssier, C.; Campos, J.; Jean-Pierre, H.; Roger, F.; Gay, B.; Carlier, J. -P.; Jumas-Bilak, E. (2009). "Negativicoccus succinicivorans gen. Nov., sp. Nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. Nov. And Acidaminococcaceae fam. Nov. In the bacterial phylum Firmicutes". International Journal of Systematic and Evolutionary Microbiology 60 (6): 1271–1279. doi:10.1099/ijs.0.013102-0. PMID 19667386.
- ^ Gladwin, Mark; Bill Trattler (2007). Clinical Microbiology made ridiculously simple. Miami, FL: MedMaster, Inc. pp. 4–5. ISBN 978-0-940780-81-1.
External links
 This article incorporates public domain material from the NCBI document "Science Primer".
This article incorporates public domain material from the NCBI document "Science Primer".
- 3D structures of proteins associated with plasma membrane of Gram-positive bacteria
- 3D structures of proteins associated with cell wall of Gram-positive bacteria
- Gram Staining Procedure and Images
Pathogenic bacteria Bacterial disease · Coley's Toxins · Exotoxin · Lysogenic cycle
Human flora Substrate preference Oxygen preference Structures Cell wall: Peptidoglycan (NAM, NAG, DAP)
Gram-positive bacteria only: Teichoic acid · Lipoteichoic acid · Endospore
Gram-negative bacteria only: Bacterial outer membrane (Porin, Lipopolysaccharide) · Periplasmic space
Mycobacteria only: Arabinogalactan · Mycolic acidOutside envelopeCompositeShapes Prokaryotes: Bacteria classification (phyla and orders) G-/
OMTerra-/Glidobacteria (BV1)Eobacteria (Chloroflexi, Deinococcus-Thermus) · Cyanobacteria · Thermodesulfobacteria · thermophiles (Aquificae · Thermotogae)Proteobacteria (BV2)BV4SphingobacteriaOther GNAcidobacteria · Chrysiogenetes · Deferribacteres · Fusobacteria · Gemmatimonadetes · Nitrospirae · Synergistetes · Dictyoglomi · LentisphaeraeG+/
no OMFirmicutes
(BV3)Actinobacteria
(BV5)Actinomycineae: Actinomycetaceae
Corynebacterineae: Mycobacteriaceae · Nocardiaceae · Corynebacteriaceae
Micrococcineae: BrevibacteriaceaeOther subclassesFirmicutes (low-G+C) Infectious diseases · Bacterial diseases: G+ (primarily A00–A79, 001–041, 080–109) Bacilli Streptococcusαoptochin susceptible: S. pneumoniae (Pneumococcal infection)optochin resistant: S. viridans: S. mitis, S. mutans, S. oralis, S. sanguinis, S. sobrinus, milleri groupβA, bacitracin susceptible: S. pyogenes (Scarlet fever, Erysipelas, Rheumatic fever, Streptococcal pharyngitis)B, bacitracin resistant, CAMP test+: S. agalactiaeungrouped: Streptococcus iniae (Cutaneous Streptococcus iniae infection)Clostridia Clostridium (spore-forming)Peptostreptococcus (non-spore forming)Peptostreptococcus magnusMollicutes MycoplasmataceaeUreaplasma urealyticum (Ureaplasma infection) · Mycoplasma genitalium · Mycoplasma pneumoniae (Mycoplasma pneumonia)Erysipelothrix rhusiopathiae (Erysipeloid)Categories:
Wikimedia Foundation. 2010.