- Hepatitis C virus
Taxobox
name = Hepatitis C virus
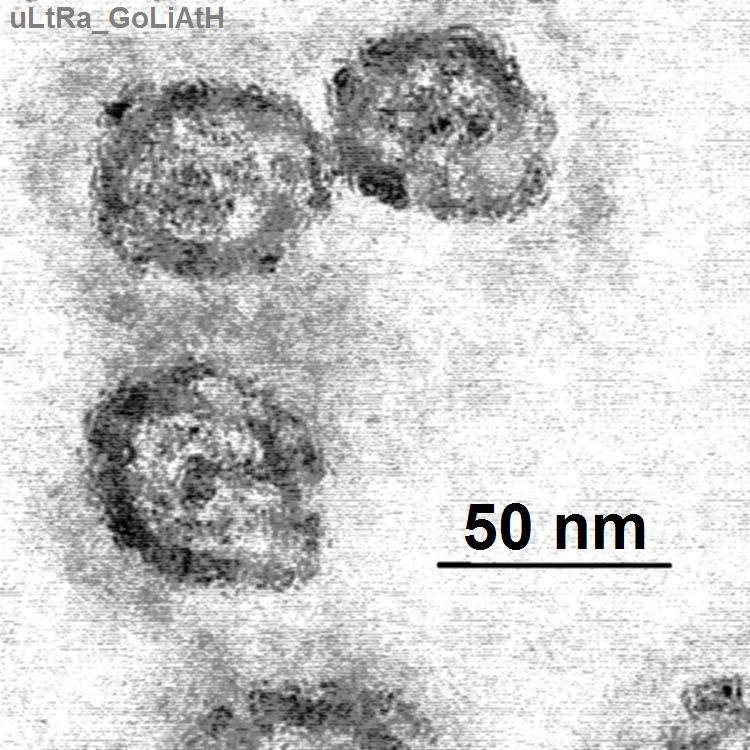
image_width = 160px
image_caption = Electron micrograph of Hepatitis C virus. The bar = 50 nm
virus_group = iv
familia =Flaviviridae
genus = "Hepacivirus "
type_species = "Hepatitis C virus":"This page is for the virus. For the disease, see
Hepatitis C ."Hepatitis C virus (HCV) is a small (50 nm in size), enveloped, positive sense single strand
RNA virus in the family "Flaviviridae ". AlthoughHepatitis A virus ,Hepatitis B virus , and Hepatitis C virus have similar names (because they all causeliver inflammation), these are distinctly different viruses both genetically and clinically.tructure
The structure of the hepatitis C virus consists of a core of genetic material (RNA), surrounded by an icosahedral protective shell of
protein , and further encased in a lipid (fatty) envelope of cellular origin. Two viral envelopeglycoprotein s, E1 and E2, are embedded in the lipid envelope.cite journal
author=Op De Beeck A, Dubuisson J
title=Topology of hepatitis C virus envelope glycoproteins
journal=Rev. Med. Virol.
volume=13
issue=4
pages=233–41
year=2003
pmid=12820185
doi=10.1002/rmv.391]Genome
Hepatitis C virus has a positive sense
RNA genome that consists of a singleopen reading frame of 9600nucleoside bases.cite journal |author=Kato N |title=Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation |journal=Microb. Comp. Genomics |volume=5 |issue=3 |pages=129–51 |year=2000 |pmid=11252351 |doi=] At the 5’ and 3’ ends of the RNA are the UTR regions, that are not translated into proteins but are important to translation and replication of the viral RNA. The 5’ UTR has aribosome binding sitecite journal |author=Jubin R |title=Hepatitis C IRES: translating translation into a therapeutic target |journal=Curr. Opin. Mol. Ther. |volume=3 |issue=3 |pages=278–87 |year=2001 |pmid=11497352 |doi=] (IRES - Internal Ribosomal Entry Site) that starts the translation of a 3000 amino acid containing protein that is later cut by cellular and viralprotease s into 10 active structural and non-structural smaller proteins.cite journal |author=Dubuisson J |title=Hepatitis C virus proteins |journal=World J. Gastroenterol. |volume=13 |issue=17 |pages=2406–15 |year=2007 |pmid=17552023 |doi=]Replication
Replication of HCV involves several steps. The viruses need a certain environment to be able to replicate, and must therefore first move to such areas.
HCV has a high rate of replication with approximately one trillion particles produced each day in an infected individual. Due to lack of proofreading by the HCV RNA polymerase, HCV also has an exceptionally high mutation rate, a factor that may help it elude the host's immune response.
HCV mainly replicates within
hepatocyte s in the liver, although there is controversial evidence for replication inlymphocyte s ormonocyte s. By mechanisms ofhost tropism , the viruses reach these proper locations. Circulating HCV particles bind to receptors on the surfaces of hepatocytes and subsequently enter the cells. Two putative HCV receptors areCD81 and humanscavenger receptor class B1 (SR-BI). However, these receptors are found throughout the body. The identification of hepatocyte-specific cofactors that determine observed HCV livertropism are currently under investigation.Once inside the hepatocyte, HCV initiates the
lytic cycle . It utilizes the intracellular machinery necessary to accomplish its own replication.cite journal | author = Lindenbach B, Rice C | title = Unravelling hepatitis C virus replication from genome to function | journal = Nature | volume = 436 | issue = 7053 | pages = 933–8 | year = 2005 | pmid = 16107832 | doi = 10.1038/nature04077] Specifically, the HCV genome is translated to produce a single protein of around 3011 amino acids. This "polyprotein" is then proteolytically processed by viral and cellularproteases to produce three structural (virion-associated) and seven nonstructural (NS) proteins. Alternatively, a frameshift may occur in the Core region to produce an Alternate Reading Frame Protein (ARFP). HCV encodes two proteases, the NS2 cysteine autoprotease and the NS3-4A serine protease. The NS proteins then recruit the viral genome into an RNA replication complex, which is associated with rearranged cytoplasmic membranes. RNA replication takes places via the viral RNA-dependentRNA polymerase of NS5B, which produces a negative-strand RNA intermediate. The negative strand RNA then serves as a template for the production of new positive-strand viral genomes. Nascent genomes can then be translated, further replicated, or packaged within new virus particles. New virus particles presumably bud into the secretory pathway and are released at the cell surface.Genotypes
Based on genetic differences between HCV isolates, the hepatitis C virus species is classified into six
genotypes (1-6) with several subtypes within each genotype (represented by letters). Subtypes are further broken down into quasispecies based on their genetic diversity. The preponderance and distribution of HCV genotypes varies globally. For example, inNorth America , genotype 1a predominates followed by 1b, 2a, 2b, and 3a. InEurope , genotype 1b is predominant followed by 2a, 2b, 2c, and 3a. Genotypes 4 and 5 are found almost exclusively inAfrica . Genotype is clinically important in determining potential response tointerferon -based therapy and the required duration of such therapy. Genotypes 1 and 4 are less responsive tointerferon -based treatment than are the other genotypes (2, 3, 5 and 6). cite journal | author = Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G, Mizokami M, Murphy D, Okamoto H, Pawlotsky J, Penin F, Sablon E, Shin-I T, Stuyver L, Thiel H, Viazov S, Weiner A, Widell A | title = Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes | journal = Hepatology | volume = 42 | issue = 4 | pages = 962–73 | year = 2005 | pmid = 16149085 | doi = 10.1002/hep.20819] Duration of standard interferon-based therapy for genotypes 1 and 4 is 48 weeks, whereas treatment for genotypes 2 and 3 is completed in 24 weeks.Infection with one genotype does not confer immunity against others, and concurrent infection with two strains is possible. In most of these cases, one of the strains removes the other from the host in a short time [cite journal | author = Laskus T, Wang LF, Radkowski M, Vargas H, Nowicki M, Wilkinson J, Rakela J. | title = Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain | journal = | volume = | issue = | pages = | year = | pmid = 11160710 | doi = ] . This finding opens the door to replace strains non-responsive to medication with others easier to treat.
Vaccination
Unlike hepatitis A and B, there is currently no
vaccine to prevent hepatitis C infection.In a 2006 study, 60 patients received four different doses of an experimental hepatitis C vaccine. All the patients produced
antibodies that the researchers believe could protect them from the virus. cite news | first=Dean | last=Edell | url=http://abclocal.go.com/kgo/story?section=edell&id=4278043 | title=Hepatitis C Vaccine Looks Promising | publisher=ABC7/KGO-TV | date =2006 | accessdate =2006-07-04] . Nevertheless, as of 2008 vaccines are still in trial process [Strickland GT, El-Kamary SS, Klenerman P, Nicosia A. Hepatitis C vaccine: supply and demand. Lancet Infect Dis. 2008 Jun;8(6):379-86.Click here to read PMID 18501853] [HCV Vaccine Development http://www.who.int/vaccine_research/diseases/viral_cancers/en/index2.html#vaccine%20development]Current Research
In 2007 the
World Community Grid launched a project where, by computer modeling of the Hepatitis C Virus (and related viruses), thousands of small molecules are screened for their potential anti-viral properties in fighting the Hepatitis C Virus. This is the first project to seek out medicines to attack the virus directly once a person is infected. This is a distributed process project similar toSETI@Home where the general public downloads the World Community Grid agent and the program (along with thousands of other users) screens thousands of molecules while their computer would be otherwise idle. If the user needs to use the computer the program sleeps. There are several different projects running, including a similar one screening for anti-AIDS drugs. The project covering the Hepatitis C Virus is called "Discovering Dengue Drugs – Together." (Dengue virus and HCV belong to the same familly, together with West Nile andYellow fever viruses [http://www.utmb.edu/discoveringdenguedrugs%2Dtogether/] ). The software and information about the project can be found at the World Community Grid web site. [http://www.worldcommunitygrid.org]Current research is focused in virus' protease inhibitors, with drugs such as Telaprevir. Barriers to the study of HCV include that fact that HCV has a very narrow host range. Hence the only animal model viable for HCV study is the
chimpanzee . The use of replicons has been very successful as well but has only been recently discovered. Finally, HCV, as with most allRNA viruses, exists as aviral quasispecies , making it very difficult to isolate a single strain or receptor type for study. [Manns MP, Foster GR, Rockstroh JK, Zeuzem S, Zoulim F, Houghton M. "The way forward in HCV treatment - finding the right path." Nat Rev Drug Discov. 2007 Dec;6(12):991-1000.]References
External links
* [http://www.liverfoundation.org American Liver Foundation: Comprehensive information about Hepatitis C, including links to chapters for finding local resources]
* [http://www.hivandhepatitis.com/hep_c/hepc_news_genotype.html Academic articles about the HCV six genotypes]
* [http://www.hepatitis-c.org Hepatitis C Virus (HCV) Details]
Wikimedia Foundation. 2010.
