- Structure and genome of HIV
-
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983.[1][2] The discovery of the virus itself was not until two years after the first major cases of AIDS associated illnesses were reported in 1981.[3][4]
Contents
Structure
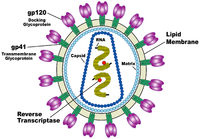
HIV is different in structure from other retroviruses. It is around 120 nm in diameter (around 60 times smaller than a red blood cell) and roughly spherical.
HIV-1 is composed of two copies of single-stranded RNA enclosed by a conical capsid comprising the viral protein p24, typical of lentiviruses (Figure 1). The RNA component is 9749 nucleotides long.[5] This is in turn surrounded by a plasma membrane of host-cell origin. The single-strand RNA is tightly bound to the nucleocapsid proteins, p6, p7 and enzymes that are indispensable for the development of the virion, such as reverse transcriptase and integrase. The nucleocapsid (p7 and p6) associates with the genomic RNA (one molecule per hexamer) and protects the RNA from digestion by nucleases. A matrix composed of an association of the viral protein p17 surrounds the capsid, ensuring the integrity of the virion particle. Also enclosed within the virion particle are Vif, Vpr, Nef, p7 and viral Protease (Figure 1). The envelope is formed when the capsid buds from the host cell, taking some of the host-cell membrane with it. The envelope includes the glycoproteins gp120 and gp41.
As a result of its role in virus-cell attachment, the structure of the virus envelope spike, consisting of gp120 and gp41, is of particular importance. It is hoped that determining the envelope spike's structure would contribute to scientific understanding of the virus and its replication cycle, and help in the creation of a cure.[6] The first model of its structure was compiled in 2006 using cryo-electron microscopy and suggested that three copies of gp120-gp41 heterodimers are thought to form a trimer as the envelope spike.[7] However, published shortly after was evidence for a single-stalk "mushroom" model, with a head consisting of a trimer gp120s and a gp41 stem, which appears as a compact structure with no obvious separation between the three monomers, anchoring it to the envelope.[8] There are various possibilities as to the source of this difference, as it is unlikely that the viruses imaged by the two groups were structurally different.[9] More recently, further evidence backing up the heterodimer trimer-based model has been found.[10]
Genome organization
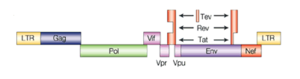
HIV has several major genes coding for structural proteins that are found in all retroviruses, and several nonstructural ("accessory") genes that are unique to HIV. The gag gene provides the basic physical infrastructure of the virus, and pol provides the basic mechanism by which retroviruses reproduce, while the others help HIV to enter the host cell and enhance its reproduction. Though they may be altered by mutation, all of these genes except tev exist in all known variants of HIV; see Genetic variability of HIV.
- gag (group-specific antigen): codes for the Gag polyprotein, which is processed during maturation to MA (matrix protein, p17); CA (capsid protein, p24); SP1 (spacer peptide 1, p2); NC (nucleocapsid protein, p7); SP2 (spacer peptide 2, p1) and p6.
- pol: codes for viral enzymes reverse transcriptase, integrase, and HIV protease.
- env (for "envelope"): codes for gp160, the precursor to gp120 and gp41, proteins embedded in the viral envelope which enable the virus to attach to and fuse with target cells.
- Transactivators: tat, rev, vpr
- tev: This gene is only present in a few HIV-1 isolates. It is a fusion of parts of the tat, env, and rev genes, and codes for a protein with some of the properties of tat, but little or none of the properties of rev.
RNA secondary structure
HIV pol-1 stem loop 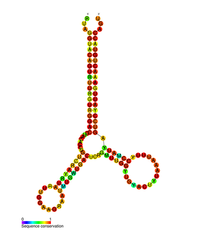
Predicted secondary structure of the HIV pol-1 stem loop Identifiers Symbol pol Rfam RF01418 Other data RNA type Cis-reg Several conserved secondary structure elements have been identified within the HIV RNA genome. These include the trans-activating responsive (TAR) element located within the 5' end of the genome and the HIV Rev response element (RRE) within the env gene.[11][12] Another RNA structure that has been identified is gag stem loop 3 (GSL3), thought to be involved in viral packaging.[13][14] RNA secondary structures have been proposed to affect the HIV life cycle by altering the function of HIV protease and reverse transcriptase, although not all elements identified have been assigned a function.
An RNA secondary structure determined by 2' hydroxyl acetylation and primer extension (SHAPE) analysis has shown to contain three stem loops and is located between the HIV protease and reverse transcriptase genes. This cis regulatory RNA has been shown to be conserved throughout the HIV family and is thought to influence the viral life cycle.[15]
The complete structure of an HIV-1 genome, extracted from infectious virions, has been solved to single-nucleotide resolution.[16]
References
- ^ Barré-Sinoussi F, Chermann JC, Rey F et al. (May 1983). "Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS)". Science 220 (4599): 868–71. Bibcode 1983Sci...220..868B. doi:10.1126/science.6189183. PMID 6189183. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=6189183.
- ^ Gallo RC, Sarin PS, Gelmann EP et al. (May 1983). "Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS)". Science 220 (4599): 865–7. doi:10.1126/science.6601823. PMID 6601823. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=6601823.
- ^ Centers for Disease Control and Prevention (1981-06-05). "Pneumocycstis Pneumonia - Los Angeles" (PDF). Morbidity and Mortality Weekly Report 30 (21): 250–2. PMID 6265753. http://www.cdc.gov/hiv/resources/reports/mmwr/pdf/mmwr05jun81.pdf. Retrieved 2008-05-10.
- ^ Centers for Disease Control and Prevention (1981-07-04). "Kaposi's Sarcoma and Pneumocycstis Pneumonia Among Homosexual Men - New York City and California" (PDF). Morbidity and Mortality Weekly Report 30 (25): 305–8. PMID 6789108. http://www.cdc.gov/hiv/resources/reports/mmwr/pdf/mmwr04jul81.pdf. Retrieved 2008-05-10.
- ^ Ratner L, Haseltine W, Patarca R et al. (1985). "Complete nucleotide sequence of the AIDS virus, HTLV-III". Nature 313 (6000): 277–84. doi:10.1038/313277a0. PMID 2578615.
- ^ "3D structure of HIV is 'revealed'". Health. BBC NEWS. 2006-01-24. http://news.bbc.co.uk/1/hi/health/4642940.stm. Retrieved 2008-08-06.
- ^ Zhu P, Liu J, Bess J Jr et al. (2006). "Distribution and three-dimensional structure of AIDS virus envelope spikes". Nature 15 (7095): 817–8. doi:10.1038/nature04817. PMID 16728975.
- ^ Zanetti G, Briggs JAG, Grunewald K et al. (2006). "Cryo-Electron Tomographic Structure of an Immunodeficiency Virus Envelope Complex In Situ". PLoS Pathology 2 (8): e83. doi:10.1371/journal.ppat.0020083. PMC 1557830. PMID 16933990. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1557830.
- ^ Sriram Subramaniam (2006). "The SIV Surface Spike Imaged by Electron Tomography: One Leg or Three?". PLoS Pathogens 2 (8): e91. doi:10.1371/journal.ppat.0020091. PMC 1557834. PMID 16933994. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1557834.
- ^ Zhu P, Winkler H, Chertova E et al. (2008). Farzan, Michael. ed. "Cryoelectron Tomography of HIV-1 Envelope Spikes: Further Evidence for Tripod-Like Legs". PLoS Pathogens 4 (11): e1000203. doi:10.1371/journal.ppat.1000203. PMC 2577619. PMID 19008954. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2577619.
- ^ Berkhout B (January 1992). "Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis". Nucleic Acids Res. 20 (1): 27–31. doi:10.1093/nar/20.1.27. PMC 310321. PMID 1738599. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=310321.
- ^ Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R (February 2002). "In vitro evidence for a long range pseudoknot in the 5'-untranslated and matrix coding regions of HIV-1 genomic RNA". J. Biol. Chem. 277 (8): 5995–6004. doi:10.1074/jbc.M108972200. PMID 11744696.
- ^ Damgaard, CK; Andersen ES, Knudsen B, Gorodkin J, Kjems J (2004). "RNA interactions in the 5' region of the HIV-1 genome". J Mol Biol 336 (2): 369–379. doi:10.1016/j.jmb.2003.12.010. PMID 14757051.
- ^ Rong, L; Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C (2003). "Deletion of stem-loop 3 is compensated by second-site mutations within the Gag protein of human immunodeficiency virus type 1". Virology 314 (1): 221–228. doi:10.1016/S0042-6822(03)00405-7. PMID 14517075.
- ^ Wang Q, Barr I, Guo F, Lee C (December 2008). "Evidence of a novel RNA secondary structurein the coding region of HIV-1 pol gene". RNA 14 (12): 2478–88. doi:10.1261/rna.1252608. PMC 2590956. PMID 18974280. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2590956.
- ^ Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM (2009). "Architecture and Secondary Structure of an Entire HIV-1 RNA Genome". Nature 460 (7256): 711–6. doi:10.1038/nature08237. PMC 2724670. PMID 19661910. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2724670.
External links
- Hunt R. "HIV and AIDS". Human Immunodeficiency Virus and AIDS. University of South Carolina School of Medicine. http://pathmicro.med.sc.edu/lecture/hiv9.htm. Retrieved 2008-08-06.
- Rfam entry for HIV pol-1 stem loop
DNA RNA capsid: matrix protein (M1 protein) · viral envelope (M2 protein)
glycoprotein: Influenza hemagglutinin · NeuraminidaseParainfluenzaParainfluenza hemagglutinin-neuraminidaseRSVRespiratory syncytial virus G proteinRT Structure and genome of HIVVSPs: gag · pol (Integrase, Reverse transcriptase, HIV-1 protease) · env (gp120, gp41)
VRAPs: transactivators (Tat, Rev, Vpr) · Nef · Vif · VpuFusion protein Categories:- HIV/AIDS
Wikimedia Foundation. 2010.