- Kringle domain
Pfam_box
Symbol = Kringle
Name =
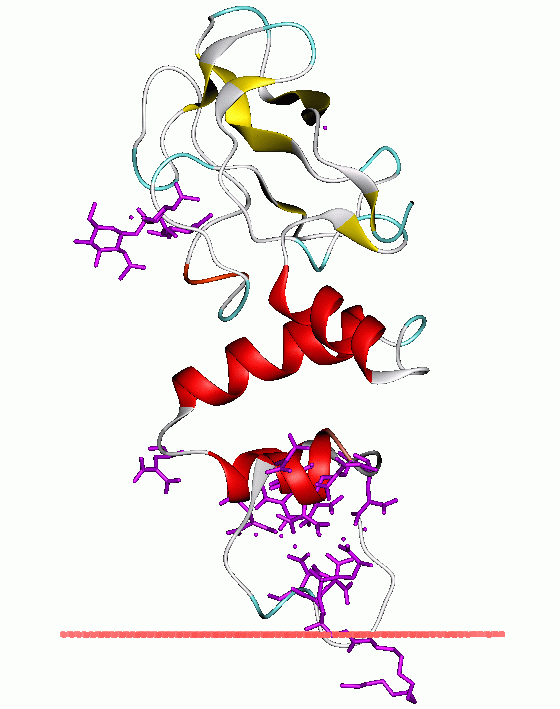
width =
caption = Bovineprothrombin fragment 1 in complex with calcium andlysophosphatidylserine
Pfam= PF00051
InterPro= IPR000001
SMART= KR
PROSITE = PDOC00020
SCOP = 1pk4
TCDB =
OPM family=
OPM protein= 1nl2
PDB=PDB3|1pmlB:215-296 PDB3|1tpkA:215-296 PDB3|1pk2 :215-296PDB3|1kdu :70-151 PDB3|1urk :70-151 PDB3|5hpgB:481-560PDB3|1ceaB:103-181 PDB3|1ki0A:103-181 PDB3|1hpk :103-181PDB3|1cebB:103-181 PDB3|1pkr :103-181 PDB3|1hpj :103-181PDB3|1krn :377-454 PDB3|1pmkB:377-454 PDB3|1pk4 :377-454PDB3|2pk4 :377-454 PDB3|1kiv :4124-4201 PDB3|3kiv :4124-4201PDB3|4kiv :4124-4201 PDB3|1jfnA:3676-3753 PDB3|1i71A:3782-3859PDB3|1i5kB:185-262 PDB3|1b2iA:185-262 PDB3|1gmoF:128-206PDB3|1bhtB:128-206 PDB3|1gp9D:128-206 PDB3|1gmnB:128-206PDB3|1nk1B:128-206 PDB3|1nl1A:109-187 PDB3|2spt :109-187PDB3|1nl2A:109-187 PDB3|2pf1 :109-187 PDB3|1a0hD:214-292PDB3|2hppP:214-292 PDB3|2hpqP:213-291Kringle Domains are autonomous
protein domain s that fold into large loops stabilized by 3 disulfide linkages. These are important inprotein -protein interactions withblood coagulation factors. The nameKringle comes from theScandinavia n pastry that these structures resemble.Kringle domains have been found in
plasminogen ,hepatocyte growth factor s,prothrombin , and apolipoprotein(a).Kringles are found throughout the
blood clotting and fibrinolytic proteins. Kringle domains are believed to play a role in binding mediators (e.g., membranes, other proteins or phospholipids), and in the regulation of proteolytic activitycite journal |author=Fujikawa K, McMullen BA |title=Amino acid sequence of the heavy chain of human alpha-factor XIIa (activated Hageman factor) |journal=J. Biol. Chem. |volume=260 |issue=9 |pages=5328–5341 |year=1985 |pmid=3886654] cite journal |author=Patthy L, Trexler M, Banyai L, Varadi A, Vali Z |title=Kringles: modules specialized for protein binding. Homology of the gelatin-binding regionof fibronectin with the kringle structures of proteases |journal=FEBS Lett. |volume=171 |issue=1 |pages=131–136 |year=1984 |pmid=6373375 |doi=10.1016/0014-5793(84)80473-1] cite journal |author=Atkinson RA, Williams RJ |title=Solution structureof the kringle 4 domain from human plasminogen by 1H nuclear magnetic resonance spectroscopy and distance geometry |journal=J. Mol. Biol. |volume=212 |issue=3 |pages=541–552 |year=1990 |pmid=2157850 |doi=10.1016/0022-2836(90)90330-O] . Kringle domainscite journal |author=Castellino FJ, Beals JM |title=The genetic relationships between the kringle domains of human plasminogen, prothrombin, tissue plasminogen activator, urokinase, and coagulation factor XII |journal=J. Mol. Evol. |volume=26 |issue=4 |pages=358–369 |year=1987 |pmid=3131537 |doi=10.1007/BF02101155] cite journal |author=Patthy L |title=Evolution of the proteases of blood coagulation and fibrinolysis by assembly from modules |journal=Cell |volume=41
issue=3 |pages=657–663 |year=1985 |pmid=3891096 |doi=10.1016/S0092-8674(85)80046-5] cite journal |author=Takahashi K, IkeoK, Gojobori T |title=Evolutionary origin of numerous kringles in human and simian apolipoprotein(a) |journal=FEBS Lett. |volume=287 |issue=1 |pages=146–148 |year=1991 |pmid=1879523 |doi=10.1016/0014-5793(91)80036-3] are characterised by a triple loop, 3-disulfide bridge structure, whose conformation is defined by a number of hydrogen bonds and small pieces of anti-parallel beta-sheet. They are found in a varying number of copies in some plasma proteins including prothrombin and urokinase-type plasminogenactivator, which are serine proteases belonging to MEROPS peptidase family S1A.Human proteins containing this domain
ATF ;F12 ;F2 ;HABP2 ;HGF ;HGFAC ;KREMEN1 ;KREMEN2 ;LPA ;LPAL2 ;MST1 ;PIK3IP1 ;PLAT ;PLAU ;PLG ;PRSS12 ;ROR1 ;ROR2 ;References
External links
* [http://www.expasy.org/cgi-bin/nicedoc.pl?PDOC00020 Kringle domain] in
PROSITE
* [http://smart.embl-heidelberg.de/smart/do_annotation.pl?DOMAIN=KR KR domain entry in the SMART database]
Wikimedia Foundation. 2010.
