- Mini-ykkC RNA motif
-
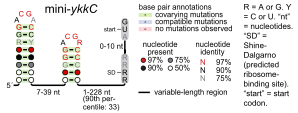 Consensus secondary structure of mini-ykkC RNAs. Layout is similar to that used in a previously published drawing.[1]
Consensus secondary structure of mini-ykkC RNAs. Layout is similar to that used in a previously published drawing.[1]mini-ykkC RNA motif 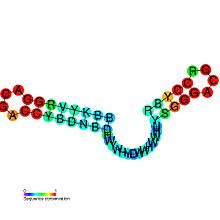
Predicted secondary structure and sequence conservation of mini-ykkC Identifiers Symbol mini-ykkC Rfam RF01068 Other data RNA type Cis-reg Domain(s) Bacteria SO 0005836 The mini-ykkC RNA motif is a putative RNA structure that is conserved in bacteria.[1] The motif consists of two conserved stem-loops whose terminal loops contain the RNA sequence ACGR, where R represents either A or G. Mini-ykkC RNAs are widespread in Proteobacteria, but some are predicted in other phyla of bacteria. It is expected that the RNAs are cis-regulatory elements, because they are typically located upstream of protein-coding genes.
The genes that are apparently controlled by mini-ykkC RNAs bear a resemblance to the genes controlled by another putative cis-regulatory RNA element called the ykkC-yxkD leader, and nine gene families are common to both. Therefore, it was proposed that these two RNA classes have the same function. However, their mechanisms might be different. While the complex structure and many conserved nucleotides found in the ykkC-yxkD leader suggest that it might be a riboswitch, these features are not present in the mini-ykkC RNA motif.
References
- ^ a b Weinberg Z, Barrick JE, Yao Z et al. (2007). "Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline". Nucleic Acids Res. 35 (14): 4809–19. doi:10.1093/nar/gkm487. PMC 1950547. PMID 17621584. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1950547.
External links
Categories:- Cis-regulatory RNA elements
- Molecular and cellular biology stubs
Wikimedia Foundation. 2010.
